5TW3
 
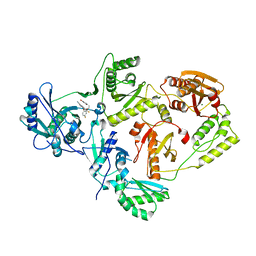 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-7-fluoro-2-naphthonitrile (JLJ636), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-7-fluoronaphthalene-2-carbonitrile, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2016-11-11 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structural and Preclinical Studies of Computationally Designed Non-Nucleoside Reverse Transcriptase Inhibitors for Treating HIV infection.
Mol. Pharmacol., 91, 2017
|
|
3MIO
 
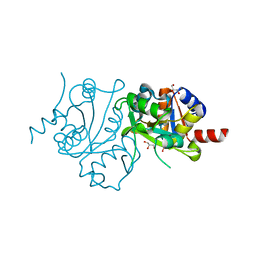 | | Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase domain from Mycobacterium tuberculosis at pH 6.00 | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Singh, M, Karthikeyan, S. | | Deposit date: | 2010-04-11 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for pH dependent monomer-dimer transition of 3,4-dihydroxy 2-butanone-4-phosphate synthase domain from Mycobacterium tuberculosis
J.Struct.Biol., 174, 2011
|
|
3MK5
 
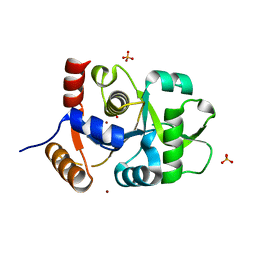 | | Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase domain from Mycobacterium tuberculosis with sulfate and zinc at pH 4.00 | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, SULFATE ION, ZINC ION | | Authors: | Singh, M, Karthikeyan, S. | | Deposit date: | 2010-04-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for pH dependent monomer-dimer transition of 3,4-dihydroxy 2-butanone-4-phosphate synthase domain from Mycobacterium tuberculosis
J.Struct.Biol., 174, 2011
|
|
6NJS
 
 | | Stat3 Core in complex with compound SD36 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-01-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
7MKV
 
 | |
6NUQ
 
 | | Stat3 Core in complex with compound SI109 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-02-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
5V2W
 
 | | Crystal structure of a LuxS from salmonella typhi | | Descriptor: | S-ribosylhomocysteine lyase, ZINC ION | | Authors: | Perumal, P, Raina, R, Manoj Kumar, P, Arockisamy, A, SundaraBaalaji, N. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a LuxS from salmonella typhi
To Be Published
|
|
8P49
 
 | |
8Q70
 
 | |
8QHP
 
 | |
8Q94
 
 | |
8Q95
 
 | | Crystal structure of the SARS-CoV-2 BA.1 RBD with neutralizing-VHHs Ma16B06 and Ma3F05 | | Descriptor: | Nanobody Ma16B06, Nanobody Ma3F05, Spike protein S1 | | Authors: | Aksu, M, Rymarenko, O, Guttler, T, Gorlich, D. | | Deposit date: | 2023-08-19 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8Q93
 
 | | Crystal structure of the SARS-COV-2 RBD with neutralizing-VHHs Re30H02 and Re21D01 | | Descriptor: | Nanobody Re21D01, Nanobody Re30H02, Spike protein S1 | | Authors: | Aksu, M, Guttler, T, Rymarenko, O, Gorlich, D. | | Deposit date: | 2023-08-19 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
4PTN
 
 | | Crystal Structure of YagE, a KDG aldolase protein in complex with Magnesium cation coordinated L-glyceraldehyde | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, L-glyceraldehyde, ... | | Authors: | Manoj Kumar, P, Baskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-03-11 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of YagE, a putative DHDPS-like protein from Escherichia coli K12.
Proteins, 71, 2008
|
|
3B6S
 
 | | Crystal Structure of hla-b*2705 Complexed with the Citrullinated Vasoactive Intestinal Peptide Type 1 Receptor (vipr) Peptide (residues 400-408) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-27 alpha chain, ... | | Authors: | Beltrami, A, Rossmann, M, Fiorillo, M.T, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, A. | | Deposit date: | 2007-10-29 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Citrullination-dependent Differential Presentation of a Self-peptide by HLA-B27 Subtypes.
J.Biol.Chem., 283, 2008
|
|
4KF7
 
 | |
4KF8
 
 | |
5DCC
 
 | | X-RAY CRYSTAL STRUCTURE OF a TEBIPENEM ADDUCT OF L,D TRANSPEPTIDASE 2 FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | (4S)-4-methyl-2,5,7-trioxoheptanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pan, Y, Basta, L, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-08-23 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase LdtMt2 by biapenem and tebipenem.
BMC Biochem., 18, 2017
|
|
5DC2
 
 | | X-RAY CRYSTAL STRUCTURE OF A ENZYMATICALLY DEGRADED BIAPENEM-ADDUCT OF L,D-TRANSPEPTIDASE 2 FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | (4S)-4-methyl-2,5,7-trioxoheptanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pan, Y, Basta, L, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-08-23 | | Release date: | 2016-10-05 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase LdtMt2 by biapenem and tebipenem.
BMC Biochem., 18, 2017
|
|
5D7H
 
 | | X-RAY CRYSTAL STRUCTURE OF L,D TRANSPEPTIDASE 2 FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | GLYCEROL, L,D-transpeptidase 2, SULFATE ION | | Authors: | Saavedra, H, Basta, L.A, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-08-13 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase LdtMt2 by biapenem and tebipenem.
BMC Biochem., 18, 2017
|
|
5AFS
 
 | | structure of Zn-bound periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, GLYCEROL, PERIPLASMIC SOLUTE BINDING PROTEIN, ... | | Authors: | Sharma, N, Selvakumar, P, Kumar, P, Sharma, A.K. | | Deposit date: | 2015-01-23 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure analysis in Zn(2+)-bound state and biophysical characterization of CLas-ZnuA2.
Biochim. Biophys. Acta, 1864, 2016
|
|
6AX5
 
 | | RPT1 region of INI1/SNF5/SMARCB1_HUMAN - SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1. | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Girvin, M.E, Cahill, S.M, Harris, R, Cowburn, D, Spira, M, Wu, X, Prakash, R, Bernowitz, M, Almo, S.C, Kalpana, G.V. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | INI1/SMARCB1 Rpt1 domain mimics TAR RNA in binding to integrase to facilitate HIV-1 replication.
Nat Commun, 12, 2021
|
|
6BMN
 
 | | Structure of human DHHC20 palmitoyltransferase, space group P63 | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, ZINC ION, ... | | Authors: | Rana, M.S, Lee, C.-J, Banerjee, A. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-24 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
6BML
 
 | | Structure of human DHHC20 palmitoyltransferase, irreversibly inhibited by 2-bromopalmitate | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, PALMITIC ACID, PHOSPHATE ION, ... | | Authors: | Rana, M.S, Lee, C.-J, Banerjee, A. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-24 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
6BMM
 
 | | Structure of human DHHC20 palmitoyltransferase, space group P21 | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,5S)-hexane-2,5-diol, PHOSPHATE ION, ... | | Authors: | Rana, M.S, Lee, C.-J, Banerjee, A. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-24 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
