7AR8
 
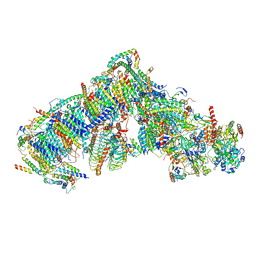 | | Cryo-EM structure of Arabidopsis thaliana complex-I (closed conformation) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | Authors: | Klusch, N, Kuelbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
6RKO
 
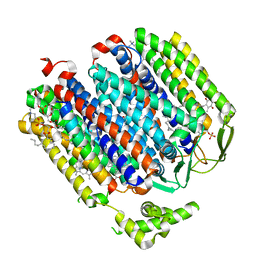 | | Cryo-EM structure of the E. coli cytochrome bd-I oxidase at 2.68 A resolution | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome bd-I ubiquinol oxidase subunit 1, ... | | Authors: | Safarian, S, Hahn, A, Kuehlbrandt, W, Michel, H. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Active site rearrangement and structural divergence in prokaryotic respiratory oxidases.
Science, 366, 2019
|
|
4CDB
 
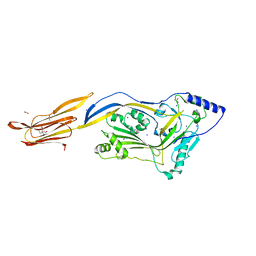 | | Crystal structure of listeriolysin O | | Descriptor: | ACETATE ION, LISTERIOLYSIN O, SODIUM ION, ... | | Authors: | Koester, S, Yildiz, O. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Listeriolysin O Reveals Molecular Details of Oligomerization and Pore Formation
Nat.Commun., 5, 2014
|
|
8R33
 
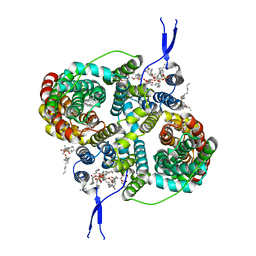 | |
8R34
 
 | | CryoEM structure of the symmetric Pho90 dimer from yeast with substrates. | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Low-affinity phosphate transporter PHO90, PHOSPHATE ION, ... | | Authors: | Schneider, S, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2023-11-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Complementary structures of the yeast phosphate transporter Pho90 provide insights into its transport mechanism.
Structure, 2024
|
|
8R35
 
 | |
7NF8
 
 | | Ovine (b0,+AT-rBAT)2 hetero-tetramer, asymmetric unit, rigid-body fitted | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, Y, Kuehlbrandt, W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-01-19 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Ca 2+ -mediated higher-order assembly of heterodimers in amino acid transport system b 0,+ biogenesis and cystinuria.
Nat Commun, 13, 2022
|
|
7NF7
 
 | | Ovine rBAT ectodomain homodimer, asymmetric unit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lee, Y, Kuehlbrandt, W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-01-19 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Ca 2+ -mediated higher-order assembly of heterodimers in amino acid transport system b 0,+ biogenesis and cystinuria.
Nat Commun, 13, 2022
|
|
7NF6
 
 | | Ovine b0,+AT-rBAT heterodimer | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, Y, Kuehlbrandt, W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-01-19 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Ca 2+ -mediated higher-order assembly of heterodimers in amino acid transport system b 0,+ biogenesis and cystinuria.
Nat Commun, 13, 2022
|
|
7ZMH
 
 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) - membrane arm | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZM7
 
 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZMG
 
 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZMB
 
 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZM8
 
 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) - membrane arm | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZME
 
 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) - membrane arm | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
8B4I
 
 | | Cryo-EM structure of the Neurospora crassa TOM core complex at 3.3 angstrom | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, DIUNDECYL PHOSPHATIDYL CHOLINE, Mitochondrial import receptor subunit Tom22, ... | | Authors: | Ornelas, P, Kuehlbrandt, W. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Two conformations of the Tom20 preprotein receptor in the TOM holo complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BEP
 
 | |
8BEF
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane core) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BEE
 
 | |
8BEL
 
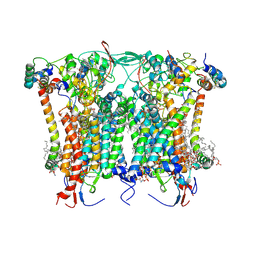 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CIII membrane domain) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BED
 
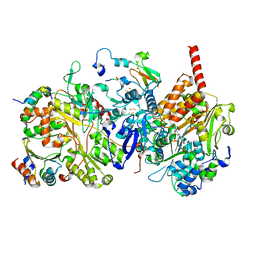 | |
8BEH
 
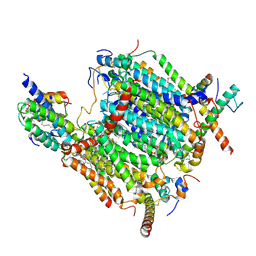 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane tip) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BPX
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete composition) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-11-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BQ5
 
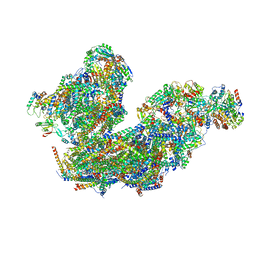 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete conformation 1 composition) | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, AT3G07480.1, Acyl carrier protein 1, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BQ6
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete conformation 2 composition) | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, AT3G07480.1, Acyl carrier protein 1, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-11-18 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
