2VLP
 
 | | R54A mutant of E9 DNase domain in complex with Im9 | | Descriptor: | COLICIN E9, COLICIN-E9 IMMUNITY PROTEIN, MALONIC ACID | | Authors: | Keeble, A.H, Joachimiak, L.A, Mate, M.J, Meenan, N, Kirkpatrick, N, Baker, D, Kleanthous, C. | | Deposit date: | 2008-01-15 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental and Computational Analyses of the Energetic Basis for Dual Recognition of Immunity Proteins by Colicin Endonucleases.
J.Mol.Biol., 379, 2008
|
|
2W8B
 
 | | Crystal structure of processed TolB in complex with Pal | | Descriptor: | ACETATE ION, GLYCEROL, PEPTIDOGLYCAN-ASSOCIATED LIPOPROTEIN, ... | | Authors: | Sharma, A, Bonsor, D.A, Kleanthous, C. | | Deposit date: | 2009-01-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Allosteric Beta-Propeller Signalling in Tolb and its Manipulation by Translocating Colicins.
Embo J., 28, 2009
|
|
2WQH
 
 | | Crystal structure of CTPR3Y3 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Krachler, A.M, Sharma, A, Kleanthous, C. | | Deposit date: | 2009-08-21 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Self-association of TPR domains: Lessons learned from a designed, consensus-based TPR oligomer.
Proteins, 78, 2010
|
|
2HQS
 
 | | Crystal structure of TolB/Pal complex | | Descriptor: | ACETATE ION, GLYCEROL, Peptidoglycan-associated lipoprotein, ... | | Authors: | Grishkovskaya, I, Bonsor, D.A, Kleanthous, C, Dodson, E.J. | | Deposit date: | 2006-07-19 | | Release date: | 2007-04-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular mimicry enables competitive recruitment by a natively disordered protein.
J.Am.Chem.Soc., 129, 2007
|
|
2GYK
 
 | | Crystal structure of the complex of the Colicin E9 DNase domain with a mutant immunity protein, IMME9 (D51A) | | Descriptor: | Colicin-E9, Colicin-E9 immunity protein, PHOSPHATE ION, ... | | Authors: | Santi, P.S, Kolade, O.O, Kuhlmann, U.C, Hemmings, A.M. | | Deposit date: | 2006-05-09 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the complexes of the Colicin E9 DNase domain with mutant immunity proteins
To be Published
|
|
6ER6
 
 | | Crystal structure of a computationally designed colicin endonuclease and immunity pair colEdes7/Imdes7 | | Descriptor: | Endonuclease colEdes7, immunity Imdes7 | | Authors: | Netzer, R, Listov, D, Dym, O, Albeck, S, Knop, O, Fleishman, S.J. | | Deposit date: | 2017-10-17 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ultrahigh specificity in a network of computationally designed protein-interaction pairs.
Nat Commun, 9, 2018
|
|
6ERE
 
 | | Crystal structure of a computationally designed colicin endonuclease and immunity pair colEdes3/Imdes3 | | Descriptor: | Immunity, PHOSPHATE ION, colicin | | Authors: | Netzer, R, Listov, D, Dym, O, Albeck, S, Knop, O, Fleishman, S.J. | | Deposit date: | 2017-10-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Ultrahigh specificity in a network of computationally designed protein-interaction pairs.
Nat Commun, 9, 2018
|
|
6FAY
 
 | | Teneurin3 monomer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Odz3 protein | | Authors: | Janssen, B.J.C, Meijer, D.H.M, van Bezouwen, L.S. | | Deposit date: | 2017-12-18 | | Release date: | 2018-03-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of Teneurin adhesion receptors reveal an ancient fold for cell-cell interaction.
Nat Commun, 9, 2018
|
|
6FB3
 
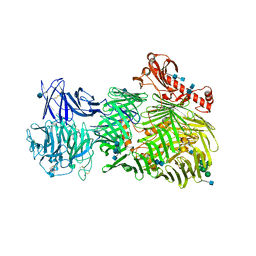 | | Teneurin 2 Partial Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Teneurin-2, ... | | Authors: | Jackson, V.A, Carrasquero, M, Lowe, E.D, Seiradake, E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-03-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structures of Teneurin adhesion receptors reveal an ancient fold for cell-cell interaction.
Nat Commun, 9, 2018
|
|
3GKL
 
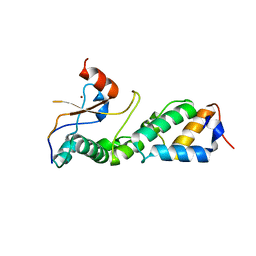 | |
3GJN
 
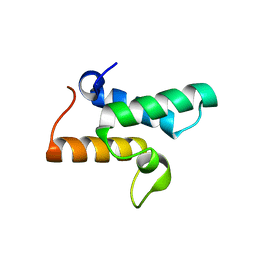 | |
2NO8
 
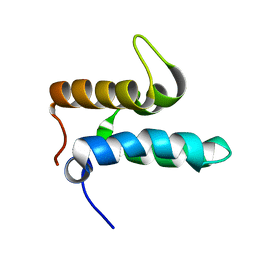 | |
4CLA
 
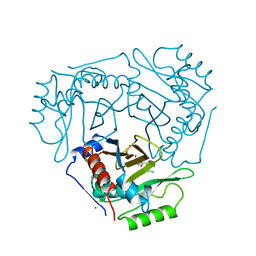 | |
1CLA
 
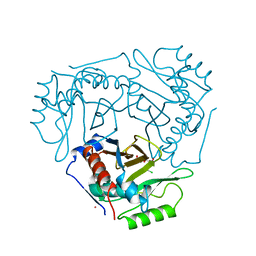 | |
2HHN
 
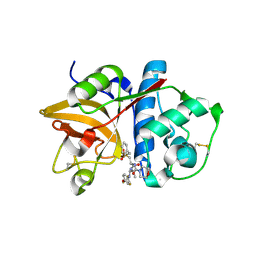 | | Cathepsin S in complex with non covalent arylaminoethyl amide. | | Descriptor: | Cathepsin S, N-[(1R)-1-[(BENZYLSULFONYL)METHYL]-2-{[(1S)-1-METHYL-2-{[4-(TRIFLUOROMETHOXY)PHENYL]AMINO}ETHYL]AMINO}-2-OXOETHYL]MORPHOLINE-4-CARBOXAMIDE, SULFATE ION | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S, Kulathila, R, Clark, K. | | Deposit date: | 2006-06-28 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synthesis and SAR of arylaminoethyl amides as noncovalent inhibitors of cathepsin S: P3 cyclic ethers
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1QFE
 
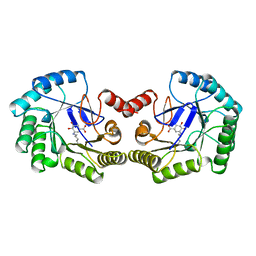 | | THE STRUCTURE OF TYPE I 3-DEHYDROQUINATE DEHYDRATASE FROM SALMONELLA TYPHI | | Descriptor: | 3-AMINO-4,5-DIHYDROXY-CYCLOHEX-1-ENECARBOXYLATE, PROTEIN (3-DEHYDROQUINATE DEHYDRATASE) | | Authors: | Shrive, A.K, Polikarpov, I, Sawyer, L, Coggins, J.R, Hawkins, A.R. | | Deposit date: | 1999-04-05 | | Release date: | 2000-04-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The two types of 3-dehydroquinase have distinct structures but catalyze the same overall reaction.
Nat.Struct.Biol., 6, 1999
|
|
3CLA
 
 | |
2CLA
 
 | |
