6JF6
 
 | |
6JFO
 
 | |
6JF3
 
 | |
6JES
 
 | |
6JEW
 
 | |
6JF9
 
 | |
6JFC
 
 | |
6JFN
 
 | |
6ILB
 
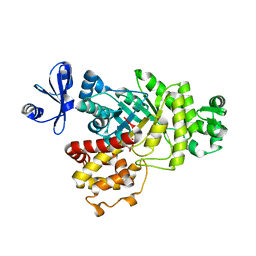 | | Native crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi | | Descriptor: | 1,2-ETHANEDIOL, Fructuronate-tagaturonate epimerase UxaE, MANGANESE (II) ION | | Authors: | Choi, M.Y, Kang, L.W, Ho, T.H, Nguyen, D.Q, Lee, I.H, Lee, J.H, Park, Y.S, Park, H.J. | | Deposit date: | 2018-10-17 | | Release date: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi
To be published
|
|
6JFD
 
 | |
6JF4
 
 | |
6JFF
 
 | |
6JFQ
 
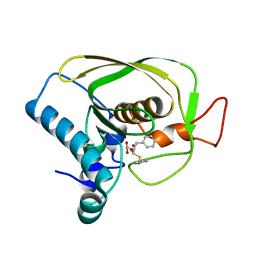 | |
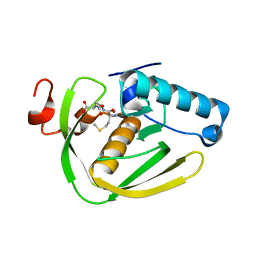
6JFA
 
 | |
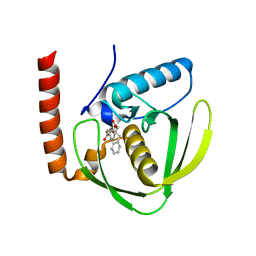
6JEU
 
 | |
6JF7
 
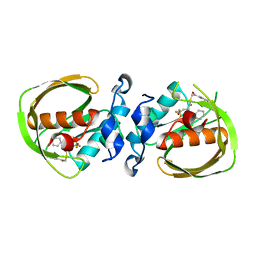 | | K3U bound crystal structure of class I type b peptide deformylase from Acinetobacter baumannii | | Descriptor: | Peptide deformylase, S-(2-oxo-2-phenylethyl) (2R)-2-benzyl-4,4,4-trifluorobutanethioate, ZINC ION | | Authors: | Jung, K.H, Ho, T.H, Lee, I.H, Kang, L.W. | | Deposit date: | 2019-02-08 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | K3U bound crystal structure of class I type b peptide deformylase from Acinetobacter baumannii
To be published
|
|
6JEX
 
 | |
6JF5
 
 | |
6JFE
 
 | |
6JEV
 
 | |
6JFG
 
 | |
3TDK
 
 | |
5XD0
 
 | | Apo Structure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glucanase, TRIETHYLENE GLYCOL | | Authors: | Baek, S.C, Ho, T.-H, Kang, L.-W, Kim, H. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Improvement of enzyme activity of beta-1,3-1,4-glucanase from Paenibacillus sp. X4 by error-prone PCR and structural insights of mutated residues.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
3PH4
 
 | | Clostridium thermocellum Ribose-5-Phosphate Isomerase B with d-allose | | Descriptor: | D-ALLOSE, Ribose-5-phosphate isomerase | | Authors: | Jung, J, Kim, J.-K, Yeom, S.-J, Ahn, Y.-J, Oh, D.-K, Kang, L.-W. | | Deposit date: | 2010-11-03 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
5I8U
 
 | | Crystal Structure of the RV1700 (MT ADPRASE) E142Q mutant | | Descriptor: | ADP-ribose pyrophosphatase, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Thirawatananond, P, Kang, L.-W, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2016-02-19 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and mutational studies of the adenosine diphosphate ribose hydrolase from Mycobacterium tuberculosis.
J. Bioenerg. Biomembr., 48, 2016
|
|
