7JM9
 
 | | Sheep Connexin-50 at 2.5 angstroms reoslution, Lipid Class 2 | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | 著者 | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | 登録日 | 2020-07-31 | | 公開日 | 2020-09-09 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JN0
 
 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 2 | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | 著者 | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | 登録日 | 2020-08-03 | | 公開日 | 2020-09-09 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | 登録日 | 1991-01-25 | | 公開日 | 1992-07-15 | | 最終更新日 | 2021-06-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
4U87
 
 | | Crystal structure of the Ba-soaked C2 crystal form of pMV158 replication initiator RepB (P3221 space group) | | 分子名称: | BARIUM ION, CHLORIDE ION, MANGANESE (II) ION, ... | | 著者 | Boer, D.R, Ruiz Maso, J.A, del Solar, G, Coll, M. | | 登録日 | 2014-08-01 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Conformational plasticity of RepB, the replication initiator protein of promiscuous streptococcal plasmid pMV158.
Sci Rep, 6, 2016
|
|
4UNR
 
 | | Mtb TMK in complex with compound 23 | | 分子名称: | 4-[3-cyano-2-oxo-7-(1H-pyrazol-4-yl)-5,6-dihydro-1H-benzo[h]quinolin-4-yl]benzoic acid, MAGNESIUM ION, Thymidylate kinase | | 著者 | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | 登録日 | 2014-05-30 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
4UNQ
 
 | | Mtb TMK in complex with compound 36 | | 分子名称: | 4-[(R)-methylsulfinyl]-2-oxo-6-[3-(trifluoromethoxy)phenyl]-1,2-dihydropyridine-3-carbonitrile, SODIUM ION, THYMIDYLATE KINASE | | 著者 | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | 登録日 | 2014-05-30 | | 公開日 | 2015-06-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
8GHS
 
 | | Empty HBV Cp183 capsid with importin-beta, subparticle reconstruction at 2-fold location | | 分子名称: | Capsid protein | | 著者 | Kim, C, Schlicksup, C.J, Hadden-Perilla, J.A, Wang, J.C.-Y, Zlotnick, A. | | 登録日 | 2023-03-10 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure of the Hepatitis B virus capsid quasi-6-fold with a trapped C-terminal domain reveals capsid movements associated with domain exit.
J.Biol.Chem., 299, 2023
|
|
8GJ8
 
 | |
8GJA
 
 | | RAD51C-XRCC3 structure | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, RAD51C, ... | | 著者 | Arvai, A.S, Tainer, J.A, Williams, G, Longo, M.A. | | 登録日 | 2023-03-15 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
4UNP
 
 | | Mtb TMK in complex with compound 34 | | 分子名称: | 5-methyl-7-propyl-1,6-naphthyridin-2(1H)-one, THYMIDYLATE KINASE | | 著者 | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | 登録日 | 2014-05-30 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
4UNN
 
 | | Mtb TMK in complex with compound 8 | | 分子名称: | 4-[3-cyano-6-(3-methoxyphenyl)-2-oxo-1H-pyridin-4-yl]benzoic acid, THYMIDYLATE KINASE | | 著者 | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | 登録日 | 2014-05-29 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
8GJ9
 
 | |
4UNS
 
 | | Mtb TMK in complex with compound 40 | | 分子名称: | N-[4-(3-CYANO-7-ETHYL-5-METHYL-2-OXO-1H-1,6-NAPHTHYRIDIN-4-YL)PHENYL]METHANESULFONAMIDE, SODIUM ION, THYMIDYLATE KINASE | | 著者 | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | 登録日 | 2014-05-30 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
8GRJ
 
 | | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone | | 分子名称: | D-glucono-1,5-lactone, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Yoshida, H, Kojima, K, Tsugawa, W, Okuda-Shimazaki, J, Kerrigan, J.A, Sode, K. | | 登録日 | 2022-09-01 | | 公開日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone
To Be Published
|
|
8IJ8
 
 | |
8IJ7
 
 | |
8IJ6
 
 | | Crystal structure of alcohol dehydrogenase from Burkholderia gladioli with NADP | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative short-chain dehydrogenases/reductase family protein | | 著者 | Han, X, Mei, Z.L, Liu, W.D, Sun, Z.T, Ma, J.A. | | 登録日 | 2023-02-26 | | 公開日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Crystal structure of alcohol dehydrogenase from Burkholderia gladioli with NADP
To Be Published
|
|
8IJG
 
 | | Crystal structure of alcohol dehydrogenase M5 from Burkholderia gladioli with NADP | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative short-chain dehydrogenases/reductase family protein | | 著者 | Han, X, Mei, Z.L, Liu, W.D, Sun, Z.T, Ma, J.A. | | 登録日 | 2023-02-27 | | 公開日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Crystal structure of alcohol dehydrogenase from Burkholderia gladioli with NADP
To Be Published
|
|
8OET
 
 | | SFX structure of the class II photolyase complexed with a thymine dimer | | 分子名称: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, DNA (14-mer), Deoxyribodipyrimidine photo-lyase, ... | | 著者 | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | 登録日 | 2023-03-12 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OF5
 
 | | Crystal structure of Aurora A 122-403 C290A, N332A, Q335A, C393A bound to ADP | | 分子名称: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Miles, J.A, Hammond, K.L.R, Bayliss, R. | | 登録日 | 2023-03-14 | | 公開日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Crystal structure of Aurora A 122-403 C290A, N332A, Q335A, C393A bound to ADP
To Be Published
|
|
8OR3
 
 | | CAND1-CUL1-RBX1-SKP1-SKP2-DCNL1 | | 分子名称: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, DCN1-like protein 1, ... | | 著者 | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | 登録日 | 2023-04-13 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
8OR0
 
 | | CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2 | | 分子名称: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, Cyclin-dependent kinase 2, ... | | 著者 | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | 登録日 | 2023-04-12 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
8OR2
 
 | | CAND1-CUL1-RBX1-DCNL1 | | 分子名称: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, DCN1-like protein 1, ... | | 著者 | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | 登録日 | 2023-04-12 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
8OR4
 
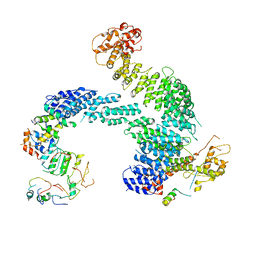 | | Partially dissociated CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2 | | 分子名称: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, Cyclin-dependent kinase 2, ... | | 著者 | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | 登録日 | 2023-04-13 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
8PI2
 
 | |
