6KPN
 
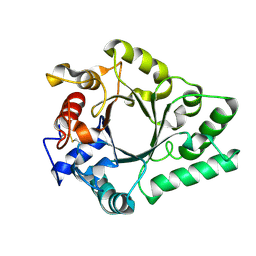 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris D154N/E156Q mutant in complex with fucosyl-N-acetylglucosamine | | Descriptor: | Chitinase, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
6KPM
 
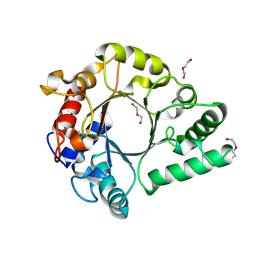 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in complex with L-fucose | | Descriptor: | Chitinase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
6KPL
 
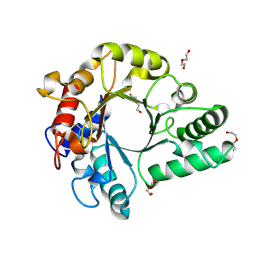 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in apo form | | Descriptor: | Chitinase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
3ABQ
 
 | |
3ABR
 
 | |
3ABO
 
 | |
3ABS
 
 | |
7OB5
 
 | |
7OBK
 
 | |
7OBX
 
 | |
7OBG
 
 | |
7OB8
 
 | |
7OBL
 
 | |
7OBC
 
 | |
7OBH
 
 | |
7OBS
 
 | |
7OBY
 
 | |
7OBD
 
 | |
7OBT
 
 | |
1MDV
 
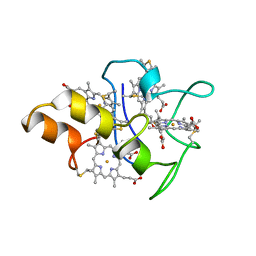 | | KEY ROLE OF PHENYLALANINE 20 IN CYTOCHROME C3: STRUCTURE, STABILITY AND FUNCTION STUDIES | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dolla, A, Arnoux, P, Protasevich, I, Lobachov, V, Brugna, M, Guidici-Orticoni, M.T, Haser, R, Czjzek, M, Makarov, A, Brushi, M. | | Deposit date: | 1998-09-08 | | Release date: | 1999-05-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Key role of phenylalanine 20 in cytochrome c3: structure, stability, and function studies.
Biochemistry, 38, 1999
|
|
6IW3
 
 | |
2D05
 
 | | Chitosanase From Bacillus circulans mutant K218P | | Descriptor: | Chitosanase, SULFATE ION | | Authors: | Fukamizo, T, Amano, S, Yamaguchi, K, Yoshikawa, T, Katsumi, T, Saito, J, Suzuki, M, Miki, K, Nagata, Y, Ando, A. | | Deposit date: | 2005-07-25 | | Release date: | 2005-12-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacillus circulans MH-K1 Chitosanase: Amino Acid Residues Responsible for Substrate Binding
J.Biochem.(Tokyo), 138, 2005
|
|
3ANY
 
 | |
3AO0
 
 | |
7XRN
 
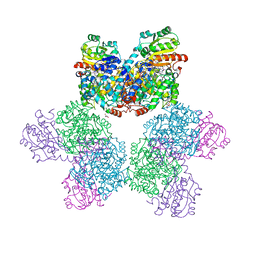 | | Ethanolamine ammonia-lyase complexed with AdoMeCbl in the presence of substrate | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-ethyl-oxolane-3,4-diol, AMMONIUM ION, COBALAMIN, ... | | Authors: | Shibata, N, Toraya, T. | | Deposit date: | 2022-05-10 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Insights into the Very Low Activity of the Homocoenzyme B 12 Adenosylmethylcobalamin in Coenzyme B 12 -Dependent Diol Dehydratase and Ethanolamine Ammonia-Lyase.
Chemistry, 28, 2022
|
|
