3ZXK
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIAXHD3, alpha-L-arabinofuranose-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3W5N
 
 | | Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase complexed with L-rhamnose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Putative rhamnosidase, ... | | Authors: | Fujimoto, Z, Jackson, A, Michikawa, M, Maehara, T, Momma, M, Henrissat, B.F, Gilbert, H.J, Kaneko, S. | | Deposit date: | 2013-01-31 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a Streptomyces avermitilis alpha-L-rhamnosidase reveals a novel carbohydrate-binding module CBM67 within the six-domain arrangement.
J.Biol.Chem., 288, 2013
|
|
3ZXL
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HIAXHD3 | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AFD
 
 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens with a partially bound cellotetraose moeity. | | Descriptor: | ENDOGLUCANASE CEL5A, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-18 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Understanding How Noncatalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan.
J.Biol.Chem., 288, 2013
|
|
4AEK
 
 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens | | Descriptor: | ENDOGLUCANASE CEL5A | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-11 | | Release date: | 2013-01-16 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan.
J.Biol.Chem., 288, 2013
|
|
2C8N
 
 | | The Structure of a family 51 arabinofuranosidase, Araf51, from Clostridium thermocellum in complex with 1,3-linked arabinoside of xylobiose. | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, alpha-L-arabinofuranose-(1-3)-alpha-D-xylopyranose | | Authors: | Taylor, E.J, Smith, N.L, Turkenburg, J.P, D'Souza, S, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-12-06 | | Release date: | 2005-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insight Into the Ligand Specificity of a Thermostable Family 51 Arabinofuranosidase, Araf51, from Clostridium Thermocellum.
Biochem.J., 395, 2006
|
|
4BA6
 
 | | High Resolution structure of the C-terminal family 65 Carbohydrate Binding Module (CBM65B) of endoglucanase Cel5A from Eubacterium cellulosolvens | | Descriptor: | Endoglucanase cel5A, GLYCEROL | | Authors: | Venditto, I, Luis, A.S, Basle, A, Temple, M, Ferreira, L.M.A, Fontes, C.M.G.A, Gilbert, H.J, Najmudin, S. | | Deposit date: | 2012-09-11 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Understanding how noncatalytic carbohydrate binding modules can display specificity for xyloglucan.
J. Biol. Chem., 288, 2013
|
|
2C7F
 
 | | The Structure of a family 51 arabinofuranosidase, Araf51, from Clostridium thermocellum in complex with 1,5-alpha-L-Arabinotriose. | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose, ... | | Authors: | Taylor, E.J, Smith, N.L, Turkenburg, J.P, D'Souza, S, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-11-23 | | Release date: | 2005-12-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight Into the Ligand Specificity of a Thermostable Family 51 Arabinofuranosidase, Araf51, from Clostridium Thermocellum.
Biochem.J., 395, 2006
|
|
1UY0
 
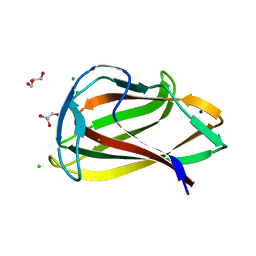 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with glc-1,3-glc-1,4-glc-1,3-glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UYX
 
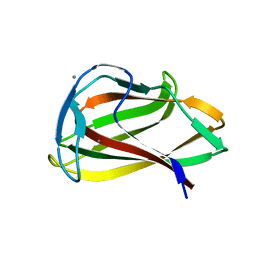 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with cellobiose | | Descriptor: | CALCIUM ION, CELLULASE B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UYZ
 
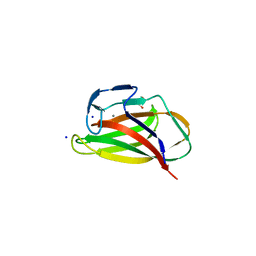 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with xylotetraose | | Descriptor: | CALCIUM ION, CELLULASE B, GLYCEROL, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UZ0
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with Glc-4Glc-3Glc-4Glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UXX
 
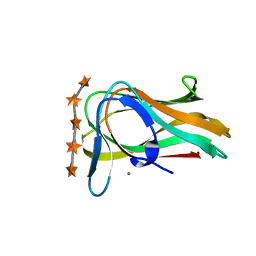 | | CBM6ct from Clostridium thermocellum in complex with xylopentaose | | Descriptor: | CALCIUM ION, XYLANASE U, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Henrissat, D.B.B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UYY
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with cellotriose | | Descriptor: | CALCIUM ION, CELLULASE B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UXZ
 
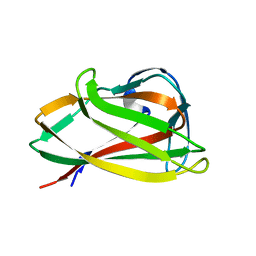 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A | | Descriptor: | CELLULASE B | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
2BGO
 
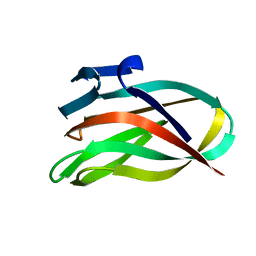 | | Mannan Binding Module from Man5C | | Descriptor: | ENDO-B1,4-MANNANASE 5C | | Authors: | Tunnicliffe, R.B, Bolam, D.N, Pell, G, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2005-01-04 | | Release date: | 2005-03-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of a Mannan-Specific Family 35 Carbohydrate-Binding Module: Evidence for Significant Conformational Changes Upon Ligand Binding
J.Mol.Biol., 347, 2005
|
|
2BM3
 
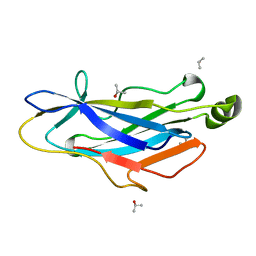 | | Structure of the Type II cohesin from Clostridium thermocellum SdbA | | Descriptor: | ISOPROPYL ALCOHOL, SCAFFOLDING DOCKERIN BINDING PROTEIN A | | Authors: | Carvalho, A.L, Gloster, T.M, Pires, V.M.R, Proctor, M.R, Prates, J.A.M, Ferreira, L.M.A, Turkenburg, J.P, Romao, M.J, Davies, G.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-03-09 | | Release date: | 2005-03-10 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights Into the Structural Determinants of Cohesin-Dockerin Specificity Revealed by the Crystal Structure of the Type II Cohesin from Clostridium Thermocellum Sdba.
J.Mol.Biol., 349, 2005
|
|
2BNJ
 
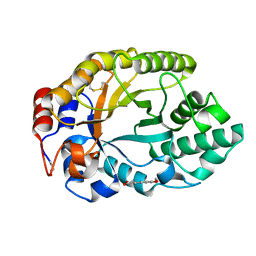 | | The xylanase TA from Thermoascus aurantiacus utilizes arabinose decorations of xylan as significant substrate specificity determinants. | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ENDO-1,4-BETA-XYLANASE, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Vardakou, M, Murray, J.W, Flint, J, Christakopoulos, P, Lewis, R.J, Gilbert, H.J. | | Deposit date: | 2005-03-25 | | Release date: | 2005-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Family 10 Thermoascus Aurantiacus Xylanase Utilizes Arabinose Decorations of Xylan as Significant Substrate Specificity Determinants.
J.Mol.Biol., 352, 2005
|
|
2C4X
 
 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-10-25 | | Release date: | 2005-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
2BO6
 
 | | DISSECTION OF MANNOSYLGLYCERATE SYNTHASE: AN ARCHETYPAL MANNOSYLTRANSFERASE | | Descriptor: | (2R)-2,3-DIHYDROXYPROPANOIC ACID, MANGANESE (II) ION, MANNOSYLGLYCERATE SYNTHASE | | Authors: | Flint, J, Taylor, E, Yang, M, Bolam, D.N, Tailford, L.E, Martinez-Fleites, C, Dodson, E.J, Davis, B.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Dissection and High-Throughput Screening of Mannosylglyceerate Synthase
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BV9
 
 | | HOW FAMILY 26 GLYCOSIDE HYDROLASES ORCHESTRATE CATALYSIS ON DIFFERENT POLYSACCHARIDES. STRUCTURE AND ACTIVITY OF A CLOSTRIDIUM THERMOCELLUM LICHENASE, CtLIC26A | | Descriptor: | ENDOGLUCANASE H | | Authors: | Taylor, E.J, Goyal, A, Guerreiro, C.I.P.D, Prates, J.A.M, Money, V.A, Ferry, N, Morland, C, Planas, A, Macdonald, J.A, Stick, R.V, Gilbert, H.J, Fontes, C.M.G.A, Davies, G.J. | | Deposit date: | 2005-06-23 | | Release date: | 2005-06-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | How Family 26 Glycoside Hydrolases Orchestrate Catalysis on Different Polysaccharides: Structure and Activity of a Clostridium Thermocellum Lichenase, Ctlic26A.
J.Biol.Chem., 280, 2005
|
|
2C24
 
 | | FAMILY 30 CARBOHYDRATE-BINDING MODULE OF CELLULOSOMAL CELLULASE CEL9D- CEL44B OF CLOSTRIDIUM THERMOCELLUM | | Descriptor: | ENDOGLUCANASE | | Authors: | Carvalho, A.L, Alves, V.D, Najmudin, S, Romao, M.J, Prates, J.A.M, Ferreira, L.M.A, Bolam, D.N, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-09-26 | | Release date: | 2005-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
2C1F
 
 | |
2BO7
 
 | | DISSECTION OF MANNOSYLGLYCERATE SYNTHASE: AN ARCHETYPAL MANNOSYLTRANSFERASE | | Descriptor: | COBALT (II) ION, GUANOSINE-5'-DIPHOSPHATE, MANNOSYLGLYCERATE SYNTHASE | | Authors: | Flint, J, Taylor, E, Yang, M, Bolam, D.N, Tailford, L.E, Martinez-Fleites, C, Dodson, E.J, Davis, B.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural dissection and high-throughput screening of mannosylglycerate synthase.
Nat. Struct. Mol. Biol., 12, 2005
|
|
2BO8
 
 | | DISSECTION OF MANNOSYLGLYCERATE SYNTHASE: AN ARCHETYPAL MANNOSYLTRANSFERASE | | Descriptor: | CHLORIDE ION, GUANOSINE 5'-(TRIHYDROGEN DIPHOSPHATE), P'-D-MANNOPYRANOSYL ESTER, ... | | Authors: | Flint, J, Taylor, E, Yang, M, Bolam, D.N, Tailford, L.E, Martinez-Fleites, C, Dodson, E.J, Davis, B.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural dissection and high-throughput screening of mannosylglycerate synthase.
Nat. Struct. Mol. Biol., 12, 2005
|
|
