7MQ3
 
 | |
7MQ1
 
 | | C9A Streptococcus pneumoniae CstR in the reduced state, space group C2 | | Descriptor: | CHLORIDE ION, Copper-sensing transcriptional repressor csoR, GLYCEROL, ... | | Authors: | Fakhoury, J.N, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2021-05-05 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Functional asymmetry and chemical reactivity of CsoR family persulfide sensors.
Nucleic Acids Res., 49, 2021
|
|
1YG4
 
 | |
1YG3
 
 | |
2AP5
 
 | |
2AP0
 
 | |
1R1V
 
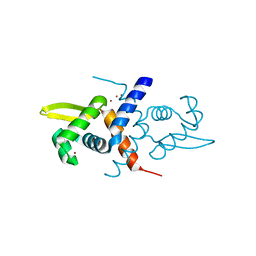 | | Crystal structure of the metal-sensing transcriptional repressor CzrA from Staphylococcus aureus in the Zn2-form | | Descriptor: | ZINC ION, repressor protein | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
1R22
 
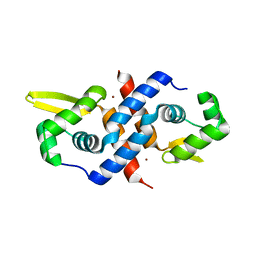 | | Crystal structure of the cyanobacterial metallothionein repressor SmtB (C14S/C61S/C121S mutant) in the Zn2alpha5-form | | Descriptor: | Transcriptional repressor smtB, ZINC ION | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
1R1U
 
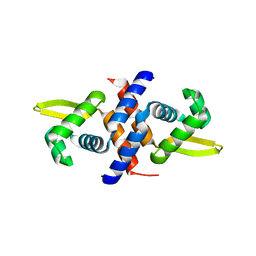 | | Crystal structure of the metal-sensing transcriptional repressor CzrA from Staphylococcus aureus in the apo-form | | Descriptor: | repressor protein | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
1R23
 
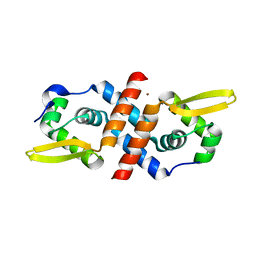 | | Crystal structure of the cyanobacterial metallothionein repressor SmtB in the Zn1-form (one Zn(II) per dimer) | | Descriptor: | Transcriptional repressor smtB, ZINC ION | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
1R1T
 
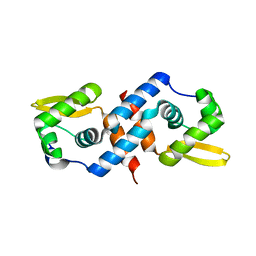 | | Crystal structure of the cyanobacterial metallothionein repressor SmtB in the apo-form | | Descriptor: | Transcriptional repressor smtB | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
3TGN
 
 | |
2KJB
 
 | |
2KJC
 
 | |
2L6I
 
 | |
2M30
 
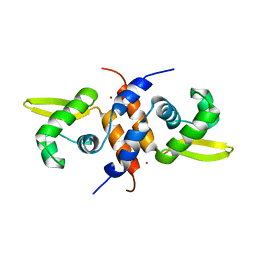 | | Solution NMR refinement of a metal ion bound protein using quantum mechanical/molecular mechanical and molecular dynamics methods | | Descriptor: | Repressor protein, ZINC ION | | Authors: | Chakravorty, D.K, Wang, B.I, Lee, C.I, Guerra, A.J, Giedroc, D.P, Merz Jr, K.M, Arunkumar, A.I, Pennella, M, Kong, X. | | Deposit date: | 2013-01-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR refinement of a metal ion bound protein using metal ion inclusive restrained molecular dynamics methods.
J.Biomol.Nmr, 56, 2013
|
|
2M0A
 
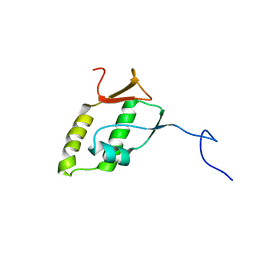 | | Solution structure of MHV nsp3a | | Descriptor: | Non-structural protein 3 | | Authors: | Keane, S.C, Giedroc, D.P. | | Deposit date: | 2012-10-23 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mouse Hepatitis Virus (MHV) nsp3a and Determinants of the Interaction with MHV Nucleocapsid (N) Protein.
J.Virol., 87, 2013
|
|
2HH7
 
 | |
6HE6
 
 | |
6HEE
 
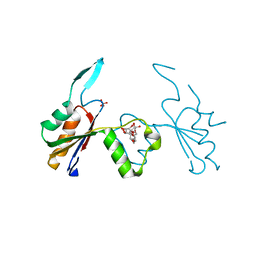 | | Crystal structure of Extracellular Domain 1 (ECD1) of FtsX from S. pneumonie in complex with undecyl-maltoside | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsX, SULFATE ION, ... | | Authors: | Martinez-Caballero, S, Alcorlo-Pages, M, Hermoso, J.A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Large Extracellular Loop of FtsX and Its Interaction with the Essential Peptidoglycan Hydrolase PcsB in Streptococcus pneumoniae.
Mbio, 10, 2019
|
|
6HFX
 
 | |
3FMS
 
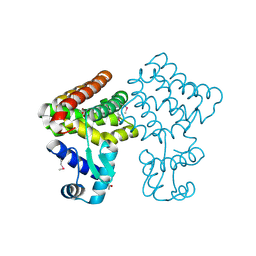 | | Crystal structure of TM0439, a GntR transcriptional regulator | | Descriptor: | ACETATE ION, NICKEL (II) ION, Transcriptional regulator, ... | | Authors: | Zheng, M, Cooper, D.R, Yu, M, Hung, L.-W, Derewenda, U, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2008-12-22 | | Release date: | 2009-02-10 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Thermotoga maritima TM0439: implications for the mechanism of bacterial GntR transcription regulators with Zn2+-binding FCD domains.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2M5P
 
 | |
2M5Q
 
 | |
2LKP
 
 | | solution structure of apo-NmtR | | Descriptor: | HTH-type transcriptional regulator NmtR | | Authors: | Lee, C, Giedroc, D. | | Deposit date: | 2011-10-18 | | Release date: | 2012-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mycobacterium tuberculosis NmtR in the Apo State: Insights into Ni(II)-Mediated Allostery.
Biochemistry, 51, 2012
|
|
