7LHJ
 
 | |
7LHM
 
 | |
7LHN
 
 | |
7LHO
 
 | |
7MS0
 
 | |
7MS3
 
 | |
7MS8
 
 | |
7MS1
 
 | |
1JKF
 
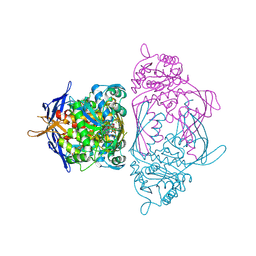 | | Holo 1L-myo-inositol-1-phosphate Synthase | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, myo-inositol-1-phosphate synthase | | 著者 | Stein, A.J, Geiger, J.H. | | 登録日 | 2001-07-12 | | 公開日 | 2002-04-10 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure and mechanism of 1-L-myo-inositol- 1-phosphate synthase
J.Biol.Chem., 277, 2002
|
|
1JKI
 
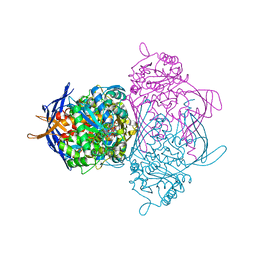 | | myo-Inositol-1-phosphate Synthase Complexed with an Inhibitor, 2-deoxy-glucitol-6-phosphate | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-DEOXY-GLUCITOL-6-PHOSPHATE, AMMONIUM ION, ... | | 著者 | Stein, A.J, Geiger, J.H. | | 登録日 | 2001-07-12 | | 公開日 | 2002-04-10 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The crystal structure and mechanism of 1-L-myo-inositol- 1-phosphate synthase
J.Biol.Chem., 277, 2002
|
|
4EFG
 
 | |
4EXZ
 
 | |
4EDE
 
 | |
4EEJ
 
 | |
4GKC
 
 | |
4ZJ0
 
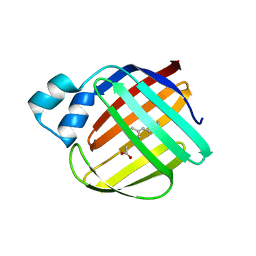 | | The crystal structure of monomer Q108K:K40L:Y60W CRBPII bound to all-trans-retinal | | 分子名称: | ACETATE ION, RETINAL, Retinol-binding protein 2 | | 著者 | Nossoni, Z, Assar, Z, Wang, W, Vasileiou, C, Borhan, B, Geiger, J.H. | | 登録日 | 2015-04-28 | | 公開日 | 2016-05-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Domain-Swapped Dimers of Intracellular Lipid-Binding Proteins: Evidence for Ordered Folding Intermediates.
Structure, 24, 2016
|
|
3CR6
 
 | |
3F8A
 
 | | Crystal Structure of the R132K:R111L:L121E:R59W Mutant of Cellular Retinoic Acid-Binding Protein Type II Complexed with C15-aldehyde (a retinal analog) at 1.95 Angstrom resolution. | | 分子名称: | 1,3,3-trimethyl-2-[(1E,3E)-3-methylpenta-1,3-dien-1-yl]cyclohexene, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular retinoic acid-binding protein 2 | | 著者 | Jia, X, Geiger, J.H. | | 登録日 | 2008-11-12 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Probing Wavelength Regulation with an Engineered Rhodopsin Mimic and a C15-Retinal Analogue
Chempluschem, 77, 2012
|
|
3FA6
 
 | |
3F9D
 
 | |
4I9S
 
 | |
4I9R
 
 | |
4LPC
 
 | |
4M6S
 
 | |
4LQ1
 
 | |
