2VEA
 
 | |
7QPK
 
 | |
1MQK
 
 | | Crystal structure of the unliganded Fv-fragment of the anti-cytochrome C oxidase antibody 7E2 | | Descriptor: | antibody 7E2 FV fragment, heavy chain, light chain | | Authors: | Essen, L.-O, Harrenga, A, Ostermeier, C, Michel, H. | | Deposit date: | 2002-09-16 | | Release date: | 2003-04-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 1.3 A X-ray structure of an antibody Fv fragment used for induced membrane-protein crystallization.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1TEZ
 
 | | COMPLEX BETWEEN DNA AND THE DNA PHOTOLYASE FROM ANACYSTIS NIDULANS | | Descriptor: | 5'-D(*AP*TP*CP*GP*GP*CP*T*(TCP)P*CP*GP*C)-3', 5'-D(*TP*CP*GP*C)-3', 5'-D(P*CP*GP*AP*AP*GP*CP*CP*GP*A)-3', ... | | Authors: | Essen, L.-O, Carell, T, Mees, A, Klar, T. | | Deposit date: | 2004-05-26 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair
Science, 306, 2004
|
|
1IVL
 
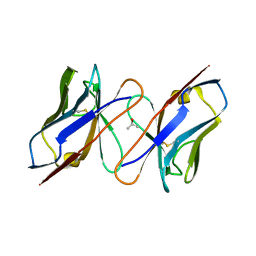 | |
1E12
 
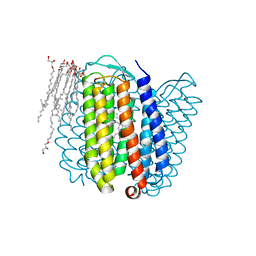 | | Halorhodopsin, a light-driven chloride pump | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, HALORHODOPSIN, ... | | Authors: | Essen, L.-O, Kolbe, M, Oesterhelt, D. | | Deposit date: | 2000-04-14 | | Release date: | 2000-06-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Light-Driven Chloride Pump Halorhodopsin at 1.8 A Resolution
Science, 288, 2000
|
|
2OG6
 
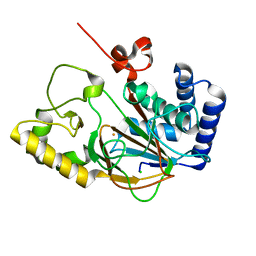 | |
2OG7
 
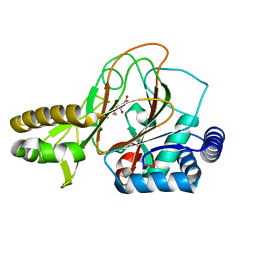 | | Cystal structure of asparagine oxygenase in complex with Fe(II), 2S,3S-3-hydroxyasparagine and succinate | | Descriptor: | BETA-HYDROXYASPARAGINE, FE (II) ION, SUCCINIC ACID, ... | | Authors: | Essen, L.-O, Strieker, M. | | Deposit date: | 2007-01-05 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mechanistic and structural basis of stereospecific Cbeta-hydroxylation in calcium-dependent antibiotic, a daptomycin-type lipopeptide
Acs Chem.Biol., 2, 2007
|
|
2VYX
 
 | |
8OV9
 
 | |
1MOG
 
 | | Crystal structure of H. salinarum dodecin | | Descriptor: | CHLORIDE ION, Dodecin, MAGNESIUM ION, ... | | Authors: | Bieger, B, Essen, L.-O, Oesterhelt, D. | | Deposit date: | 2002-09-09 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Halophilic Dodecin: A Novel, Dodecameric Flavin Binding Protein from Halobacterium salinarum
Structure, 11, 2003
|
|
8OV8
 
 | | Crystal structure of Ene-reductase 1 from black poplar mushroom in complex to NADP | | Descriptor: | Ene-reductase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Korf, L, Essen, L.-O, Karrer, D, Ruehl, M. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Shifting the substrate scope of an ene/yne-reductase by loop engineering
To Be Published
|
|
7ZHF
 
 | |
7ZHK
 
 | |
2CBG
 
 | |
2CB9
 
 | |
5A8J
 
 | | Crystal structure of the ArnB paralog VWA2 from Sulfolobus acidocaldarius | | Descriptor: | CHLORIDE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Hoffmann, L, Anders, K, Reimann, J, Linne, U, Essen, L.-O, Albers, S.-V. | | Deposit date: | 2015-07-16 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Phosphorylation-Dependent Interaction between an Archaeal Von Willebrand and Fha Domain Recruits Arna-Arnb Complex to DNA for Repression of Motility
To be Published
|
|
5A3L
 
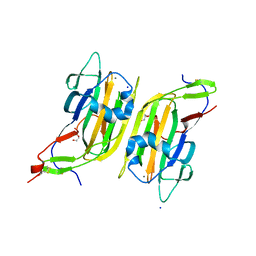 | | Structure of Cea1A in complex with N-Acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kock, M, Brueckner, S, Wozniak, N, Veelders, M, Schlereth, J, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2015-06-02 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | High-Affinity Recognition of Non-Reducing Chitinous Ends by the Yeast Adhesin Cea1
To be Published
|
|
5A3M
 
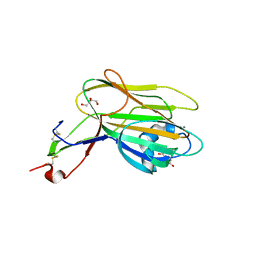 | | Structure of Cea1A in complex with Chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CEA1, ... | | Authors: | Kock, M, Brueckner, S, Wozniak, N, Veelders, M, Schlereth, J, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2015-06-02 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-Affinity Recognition of Non-Reducing Chitinous Ends by the Yeast Adhesin Cea1
To be Published
|
|
5A8I
 
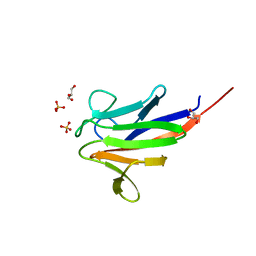 | | Crystal structure of the FHA domain of ArnA from Sulfolobus acidocaldarius | | Descriptor: | ARNA, GLYCEROL, SULFATE ION | | Authors: | Hoffmann, L, Anders, K, Reimann, J, Linne, U, Essen, L.-O, Albers, S.-V. | | Deposit date: | 2015-07-16 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phosphorylation-Dependent Interaction between an Archaeal Von Willebrand and Fha Domain Recruits Arna-Arnb Complex to DNA for Repression of Motility
To be Published
|
|
1E0R
 
 | | Beta-apical domain of thermosome | | Descriptor: | THERMOSOME | | Authors: | Bosch, G, Baumeister, W, Essen, L.-O. | | Deposit date: | 2000-04-06 | | Release date: | 2000-08-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Beta-Apical Domain from Thermosome Reveals Structural Plasticity in Protrusion Region
J.Mol.Biol., 301, 2000
|
|
3GF7
 
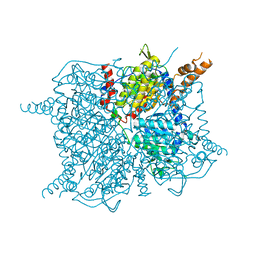 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum apoprotein | | Descriptor: | Glutaconyl-CoA decarboxylase subunit A, SULFATE ION | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-02-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GF3
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with glutaconyl-coA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Glutaconyl-CoA decarboxylase subunit A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-02-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GMA
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with glutaryl-CoA | | Descriptor: | Glutaconyl-CoA decarboxylase subunit A, glutaryl-coenzyme A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-03-13 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GLM
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with crotonyl-coA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Glutaconyl-CoA decarboxylase subunit A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-03-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
