1QD5
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI | | Descriptor: | OUTER MEMBRANE PHOSPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Ubarretxena-Belandia, I, Blaauw, M, Kalk, K.H, Verheij, H.M, Egmond, M.R, Dekker, N, Dijkstra, B.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural evidence for dimerization-regulated activation of an integral membrane phospholipase.
Nature, 401, 1999
|
|
1H1I
 
 | | CRYSTAL STRUCTURE OF QUERCETIN 2,3-DIOXYGENASE ANAEROBICALLY COMPLEXED WITH THE SUBSTRATE QUERCETN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2002-07-15 | | Release date: | 2002-11-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Anaerobic Enzyme.Substrate Structures Provide Insight Into the Reaction Mechanism of the Copper- Dependent Quercetin 2,3-Dioxygenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GUK
 
 | |
2DHE
 
 | |
1QD6
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI | | Descriptor: | 1-HEXADECANOSULFONIC ACID, CALCIUM ION, OUTER MEMBRANE PHOSPHOLIPASE (OMPLA), ... | | Authors: | Snijder, H.J, Ubarretxena-Belandia, I, Blaauw, M, Kalk, K.H, Verheij, H.M, Egmond, M.R, Dekker, N, Dijkstra, B.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evidence for dimerization-regulated activation of an integral membrane phospholipase.
Nature, 401, 1999
|
|
2DHD
 
 | |
2DHC
 
 | |
2EDC
 
 | |
2EDA
 
 | |
3TTO
 
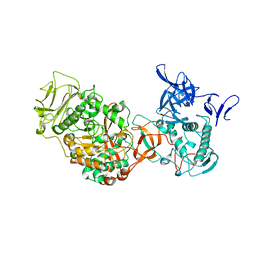 | | Crystal structure of Leuconostoc mesenteroides NRRL B-1299 N-terminally truncated dextransucrase DSR-E in triclinic form | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL | | Authors: | Brison, Y, Pijning, T, Fabre, E, Mourey, L, Morel, S, Potocki-Veronese, G, Monsan, P, Tranier, S, Remaud-Simeon, M, Dijkstra, B.W. | | Deposit date: | 2011-09-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Functional and structural characterization of alpha-(1-2) branching sucrase derived from DSR-E glucansucrase
J.Biol.Chem., 287, 2012
|
|
3TTQ
 
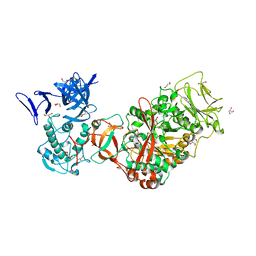 | | Crystal structure of Leuconostoc mesenteroides NRRL B-1299 N-terminally truncated dextransucrase DSR-E in orthorhombic apo-form at 1.9 angstrom resolution | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL, ... | | Authors: | Brison, Y, Pijning, T, Fabre, E, Mourey, L, Morel, S, Potocki-Veronese, G, Monsan, P, Remaud-Simeon, M, Dijkstra, B.W, Tranier, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and structural characterization of alpha-(1-2) branching sucrase derived from DSR-E glucansucrase
J.Biol.Chem., 287, 2012
|
|
4R9L
 
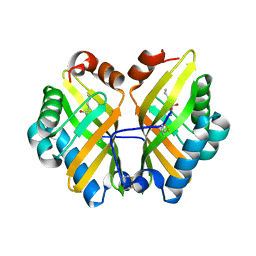 | | Structure of a thermostable elevenfold mutant of limonene epoxide hydrolase from Rhodococcus erythropolis, containing two stabilizing disulfide bonds | | Descriptor: | (2R)-2-hydroxyhexanamide, Limonene-1,2-epoxide hydrolase | | Authors: | Floor, R.J, Wijma, H.J, Jekel, P.A, Terwisscha van Scheltinga, A.C, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic validation of structure predictions used in computational design for protein stabilization.
Proteins, 83, 2015
|
|
5EHA
 
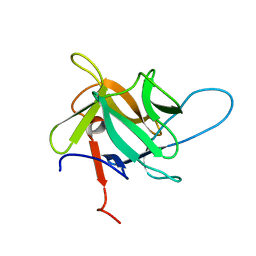 | | Crystal structure of recombinant MtaL at 1.35 Angstrom resolution | | Descriptor: | Lectin-like fold protein | | Authors: | Lai, X.-L, Soler-Lopez, M, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of recombinant tyrosinase-binding protein MtaL at 1.35 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4R9K
 
 | | Structure of thermostable eightfold mutant of limonene epoxide hydrolase from Rhodococcus erythropolis | | Descriptor: | (2R)-2-hydroxyhexanamide, GLYCEROL, Limonene-1,2-epoxide hydrolase | | Authors: | Floor, R.J, Wijma, H.J, Jekel, P.A, Terwisscha van Scheltinga, A.C, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic validation of structure predictions used in computational design for protein stabilization.
Proteins, 83, 2015
|
|
5G09
 
 | | The crystal structure of a S-selective transaminase from Bacillus megaterium bound with R-alpha-methylbenzylamine | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | Authors: | van Oosterwijk, N, Willies, S, Hekelaar, J, Terwisscha van Scheltinga, A.C, Turner, N.J, Dijkstra, B.W. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Substrate Range and Enantioselectivity of Two S-Selective Omega- Transaminases
Biochemistry, 55, 2016
|
|
4V2Q
 
 | | Ironing out their differences: Dissecting the structural determinants of a phenylalanine aminomutase and ammonia lyase | | Descriptor: | PHENYLALANINE AMMONIA-LYASE | | Authors: | Heberling, M, Masman, M, Bartsch, S, Wybenga, G.G, Dijkstra, B.W, Marrink, S, Janssen, D. | | Deposit date: | 2014-10-14 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ironing Out Their Differences: Dissecting the Structural Determinants of a Phenylalanine Aminomutase and Ammonia Lyase.
Acs Chem.Biol., 10, 2015
|
|
1B6G
 
 | | HALOALKANE DEHALOGENASE AT PH 5.0 CONTAINING CHLORIDE | | Descriptor: | CHLORIDE ION, GLYCEROL, HALOALKANE DEHALOGENASE, ... | | Authors: | Ridder, I.S, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-01-14 | | Release date: | 1999-07-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Haloalkane dehalogenase from Xanthobacter autotrophicus GJ10 refined at 1.15 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1TCM
 
 | |
1BEZ
 
 | | HALOALKANE DEHALOGENASE MUTANT WITH TRP 175 REPLACED BY TYR AT PH 5 | | Descriptor: | ACETIC ACID, HALOALKANE DEHALOGENASE | | Authors: | Ridder, I.S, Vos, G.J, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1998-05-18 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic analysis and X-ray structure of haloalkane dehalogenase with a modified halide-binding site.
Biochemistry, 37, 1998
|
|
1BE0
 
 | | HALOALKANE DEHALOGENASE AT PH 5.0 CONTAINING ACETIC ACID | | Descriptor: | ACETATE ION, ACETIC ACID, HALOALKANE DEHALOGENASE | | Authors: | Ridder, I.S, Vos, G.J, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1998-05-18 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Kinetic analysis and X-ray structure of haloalkane dehalogenase with a modified halide-binding site.
Biochemistry, 37, 1998
|
|
1BEE
 
 | | HALOALKANE DEHALOGENASE MUTANT WITH TRP 175 REPLACED BY TYR | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Ridder, I.S, Vos, G.J, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1998-05-13 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Kinetic analysis and X-ray structure of haloalkane dehalogenase with a modified halide-binding site.
Biochemistry, 37, 1998
|
|
3P0B
 
 | |
3QZU
 
 | | Crystal structure of Bacillus subtilis Lipase A 7-fold mutant; the outcome of directed evolution towards thermostability | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipase estA, ... | | Authors: | Pijning, T, Augustyniak, W, Reetz, M.T, Dijkstra, B.W. | | Deposit date: | 2011-03-07 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biophysical characterization of mutants of Bacillus subtilis lipase evolved for thermostability: Factors contributing to increased activity retention.
Protein Sci., 21, 2012
|
|
2HAD
 
 | |
3H8G
 
 | | Bestatin complex structure of leucine aminopeptidase from Pseudomonas putida | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, BICARBONATE ION, Cytosol aminopeptidase, ... | | Authors: | Kale, A, Dijkstra, B.W, Sonke, T, Thunnissen, A.M.W.H. | | Deposit date: | 2009-04-29 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the leucine aminopeptidase from Pseudomonas putida reveals the molecular basis for its enantioselectivity and broad substrate specificity.
J.Mol.Biol., 398, 2010
|
|
