2HZ8
 
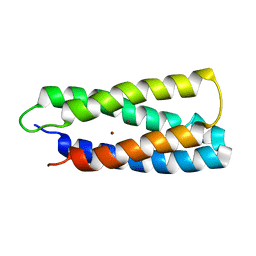 | | QM/MM structure refined from NMR-structure of a single chain diiron protein | | Descriptor: | De novo designed diiron protein, ZINC ION | | Authors: | Calhoun, J.R, Liu, W, Spiegel, K, Dal Peraro, M, Klein, M.L, Wand, A.J, DeGrado, W.F. | | Deposit date: | 2006-08-08 | | Release date: | 2007-07-17 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a designed metalloprotein and complementary molecular dynamics refinement.
Structure, 16, 2008
|
|
1Y47
 
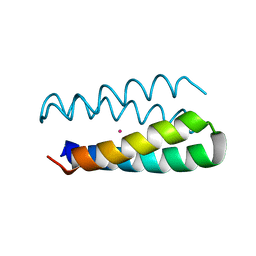 | | Structural studies of designed alpha-helical hairpins | | Descriptor: | CADMIUM ION, dueferri (DF2) | | Authors: | Lahr, S.J, Engel, D.E, Stayrook, S.E, Maglio, O, North, B, Geremia, S, Lombardi, A, DeGrado, W.F. | | Deposit date: | 2004-11-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis and design of turns in alpha-helical hairpins
J.Mol.Biol., 346, 2005
|
|
1YOD
 
 | |
2A3D
 
 | | SOLUTION STRUCTURE OF A DE NOVO DESIGNED SINGLE CHAIN THREE-HELIX BUNDLE (A3D) | | Descriptor: | PROTEIN (DE NOVO THREE-HELIX BUNDLE) | | Authors: | Walsh, S.T.R, Cheng, H, Bryson, J.W, Roder, H, Degrado, W.F. | | Deposit date: | 1999-04-01 | | Release date: | 1999-05-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of a de novo designed three-helix bundle protein.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1AL1
 
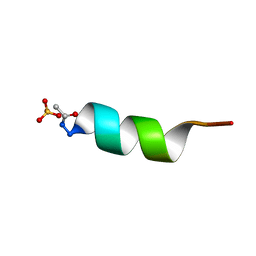 | | CRYSTAL STRUCTURE OF ALPHA1: IMPLICATIONS FOR PROTEIN DESIGN | | Descriptor: | ALPHA HELIX PEPTIDE: ELLKKLLEELKG, SULFATE ION | | Authors: | Hill, C.P, Anderson, D.H, Wesson, L, Degrado, W.F, Eisenberg, D. | | Deposit date: | 1990-07-02 | | Release date: | 1991-10-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of alpha 1: implications for protein design.
Science, 249, 1990
|
|
1COI
 
 | | DESIGNED TRIMERIC COILED COIL-VALD | | Descriptor: | COIL-VALD, SULFATE ION | | Authors: | Ogihara, N.L, Weiss, M.S, Degrado, W.F, Eisenberg, D. | | Deposit date: | 1996-08-10 | | Release date: | 1997-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the designed trimeric coiled coil coil-VaLd: implications for engineering crystals and supramolecular assemblies.
Protein Sci., 6, 1997
|
|
6YB1
 
 | | Crystal structure of an antiparallel octameric transmembrane coiled coil K2-CCTM-VbIc | | Descriptor: | GLYCINE, K2-CCTM-VbIc, NONAETHYLENE GLYCOL | | Authors: | Kratochvil, H.T, Liu, L, Scott, A.J, Woolfson, D.N, DeGrado, W.F. | | Deposit date: | 2020-03-15 | | Release date: | 2021-04-07 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Constructing ion channels from water-soluble alpha-helical barrels.
Nat.Chem., 13, 2021
|
|
1NVO
 
 | | Solution structure of a four-helix bundle model, apo-DF1 | | Descriptor: | Homodimeric Alpha2 Four-Helix Bundle | | Authors: | Maglio, O, Nastri, F, Pavone, V, Lombardi, A, DeGrado, W.F. | | Deposit date: | 2003-02-04 | | Release date: | 2003-03-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Preorganization of molecular binding sites in designed diiron proteins
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3LBW
 
 | | High resolution crystal structure of transmembrane domain of M2 | | Descriptor: | 4-bromobenzoic acid, DI(HYDROXYETHYL)ETHER, M2 protein, ... | | Authors: | Acharya, R, Polishchuk, A.L, DeGrado, W.F. | | Deposit date: | 2010-01-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3S0R
 
 | |
5ET3
 
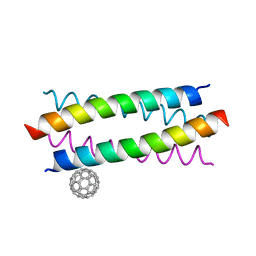 | | Crystal Structure of De novo Designed Fullerene organizing peptide | | Descriptor: | (C_{60}-I_{h})[5,6]fullerene, Fullerene Organizing Protein (C60Sol-COP-3) | | Authors: | Kim, K.-H, Kim, Y.H, Acharya, R, Kim, N.H, Paul, J, Grigoryan, G, DeGrado, W.F. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.671 Å) | | Cite: | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
2KQT
 
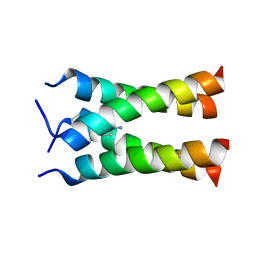 | | Solid-state NMR structure of the M2 transmembrane peptide of the influenza A virus in DMPC lipid bilayers bound to deuterated amantadine | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, M2 protein | | Authors: | Cady, S.D, Schmidt-Rohr, K, Wang, J, Soto, C.S, DeGrado, W.F, Hong, M. | | Deposit date: | 2009-11-18 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-08 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers
Nature, 463, 2010
|
|
2MUZ
 
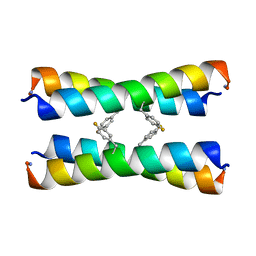 | | ssNMR structure of a designed rocker protein | | Descriptor: | designed rocker protein | | Authors: | Wang, T, Joh, N, Wu, Y, DeGrado, W.F, Hong, M. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2015-01-14 | | Method: | SOLUTION NMR | | Cite: | De novo design of a transmembrane Zn2+-transporting four-helix bundle.
Science, 346, 2014
|
|
6MCT
 
 | |
6MJH
 
 | |
6MQU
 
 | |
6MQ2
 
 | |
6MPW
 
 | |
6NV1
 
 | | Structure of drug-resistant V27A mutant of the influenza M2 proton channel bound to spiroadamantyl amine inhibitor | | Descriptor: | (1r,1'S,3'S,5'S,7'S)-spiro[cyclohexane-1,2'-tricyclo[3.3.1.1~3,7~]decan]-4-amine, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, ... | | Authors: | Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2019-02-04 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray Crystal Structures of the Influenza M2 Proton Channel Drug-Resistant V27A Mutant Bound to a Spiro-Adamantyl Amine Inhibitor Reveal the Mechanism of Adamantane Resistance.
Biochemistry, 59, 2020
|
|
6O3N
 
 | |
6OUG
 
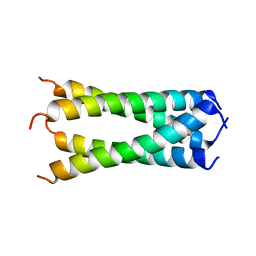 | | Structure of drug-resistant V27A mutant of the influenza M2 proton channel bound to spiroadamantyl amine inhibitor, TM + cytosolic helix construct | | Descriptor: | (1r,1'S,3'S,5'S,7'S)-spiro[cyclohexane-1,2'-tricyclo[3.3.1.1~3,7~]decan]-4-amine, Matrix protein 2 | | Authors: | Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2019-05-04 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | X-ray Crystal Structures of the Influenza M2 Proton Channel Drug-Resistant V27A Mutant Bound to a Spiro-Adamantyl Amine Inhibitor Reveal the Mechanism of Adamantane Resistance.
Biochemistry, 59, 2020
|
|
5WOC
 
 | |
6B17
 
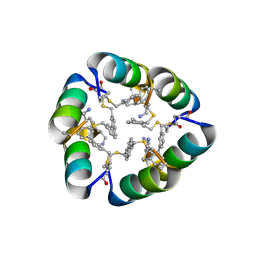 | | Design of a short thermally stable alpha-helix embedded in a macrocycle | | Descriptor: | 3,3'-dimethyl-1,1'-biphenyl, Capped-strapped peptide | | Authors: | Wu, H, Acharyya, A, Wu, Y, Liu, L, Jo, H, Gai, F, DeGrado, W.F. | | Deposit date: | 2017-09-17 | | Release date: | 2018-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design of a Short Thermally Stable alpha-Helix Embedded in a Macrocycle.
Chembiochem, 19, 2018
|
|
6ANF
 
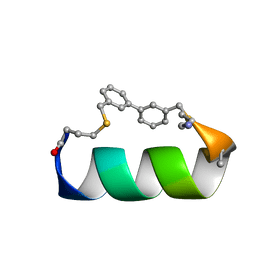 | | Design of a short thermo-stable alpha-helix embedded in a macrocycle | | Descriptor: | 3,3'-dimethyl-1,1'-biphenyl, Capped-strapped peptide | | Authors: | Wu, H, Acharyya, A, Wu, Y, Liu, L, Jo, H, Gai, F, DeGrado, W.F. | | Deposit date: | 2017-08-13 | | Release date: | 2018-02-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Design of a Short Thermally Stable alpha-Helix Embedded in a Macrocycle.
Chembiochem, 19, 2018
|
|
6BKL
 
 | | Influenza A M2 transmembrane domain bound to rimantadine | | Descriptor: | (1S)-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]ethan-1-amine, Matrix protein 2, RIMANTADINE | | Authors: | Thomaston, J.L, DeGrado, W.F. | | Deposit date: | 2017-11-08 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Inhibitors of the M2 Proton Channel Engage and Disrupt Transmembrane Networks of Hydrogen-Bonded Waters.
J. Am. Chem. Soc., 140, 2018
|
|
