4Q42
 
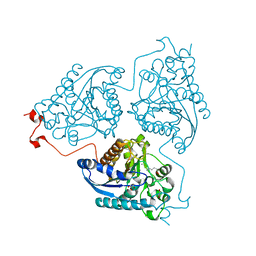 | | Crystal structure of Schistosoma mansoni arginase in complex with L-ornithine | | Descriptor: | Arginase, GLYCEROL, L-ornithine, ... | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
4RHI
 
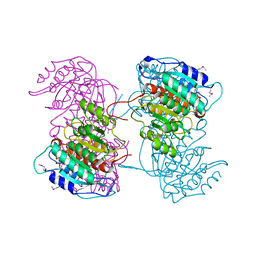 | |
4RHK
 
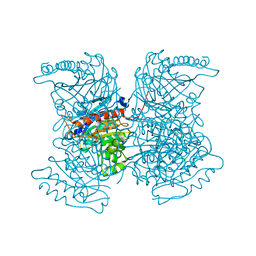 | |
4RHL
 
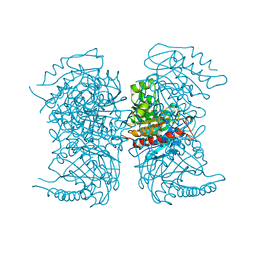 | |
4RHQ
 
 | | Crystal structure of T. brucei arginase-like protein double mutant S149D/S153D | | Descriptor: | 1,2-ETHANEDIOL, Arginase, GLYCEROL | | Authors: | Hai, Y, Barrett, M.P, Christianson, D.W. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of an Arginase-like Protein from Trypanosoma brucei That Evolved without a Binuclear Manganese Cluster.
Biochemistry, 54, 2015
|
|
4RHJ
 
 | |
4RHM
 
 | | Crystal structure of T. brucei arginase-like protein quadruple mutant S149D/R151H/S153D/S226D | | Descriptor: | 1,2-ETHANEDIOL, Arginase, GLYCEROL, ... | | Authors: | Hai, Y, Barrett, M.P, Christianson, D.W. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Arginase-like Protein from Trypanosoma brucei That Evolved without a Binuclear Manganese Cluster.
Biochemistry, 54, 2015
|
|
4RLA
 
 | |
4LA6
 
 | |
4LA5
 
 | |
1URT
 
 | | MURINE CARBONIC ANHYDRASE V | | Descriptor: | CARBONIC ANHYDRASE V, ZINC ION | | Authors: | Boriack-Sjodin, P.A, Christianson, D.W. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of an intramolecular proton transfer site in murine carbonic anhydrase V.
Biochemistry, 35, 1996
|
|
1WVA
 
 | | Crystal structure of human arginase I from twinned crystal | | Descriptor: | Arginase 1, MANGANESE (II) ION, S-2-(BORONOETHYL)-L-CYSTEINE | | Authors: | Di Costanzo, L, Sabio, G, Mora, A, Rodriguez, P.C, Ochoa, A.C, Centeno, F, Christianson, D.W. | | Deposit date: | 2004-12-14 | | Release date: | 2005-09-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in the immune response
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1PS1
 
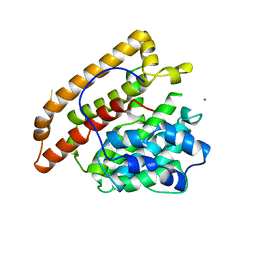 | | PENTALENENE SYNTHASE | | Descriptor: | PENTALENENE SYNTHASE, TRIMETHYL LEAD ION | | Authors: | Lesburg, C.A, Christianson, D.W. | | Deposit date: | 1997-03-23 | | Release date: | 1998-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology.
Science, 277, 1997
|
|
1RLA
 
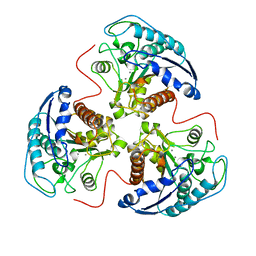 | | THREE-DIMENSIONAL STRUCTURE OF RAT LIVER ARGINASE, THE BINUCLEAR MANGANESE METALLOENZYME OF THE UREA CYCLE | | Descriptor: | ARGINASE, MANGANESE (II) ION | | Authors: | Kanyo, Z, Scolnick, L, Ash, D, Christianson, D.W. | | Deposit date: | 1996-08-15 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a unique binuclear manganese cluster in arginase.
Nature, 383, 1996
|
|
7SGI
 
 | |
7SGK
 
 | |
7SGG
 
 | |
7SGJ
 
 | |
4FCI
 
 | |
4FCK
 
 | |
7U69
 
 | |
7U6A
 
 | |
7U6B
 
 | |
7U3M
 
 | |
4GWC
 
 | | Crystal Structure of Mn2+2,Zn2+-Human Arginase I | | Descriptor: | Arginase-1, MANGANESE (II) ION, ZINC ION | | Authors: | D'Antonio, E.L, Hai, Y, Christianson, D.W. | | Deposit date: | 2012-09-01 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of non-native metal clusters in human arginase I.
Biochemistry, 51, 2012
|
|
