2A9M
 
 | |
2A9N
 
 | | A Mutation Designed to Alter Crystal Packing Permits Structural Analysis of a Tight-binding Fluorescein-scFv complex | | Descriptor: | 4-(2,7-DIFLUORO-6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)ISOPHTHALIC ACID, fluorescein-scfv | | Authors: | Cambillau, C, Spinelli, S, Honegger, A, Pluckthun, A. | | Deposit date: | 2005-07-12 | | Release date: | 2005-10-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A mutation designed to alter crystal packing permits structural analysis of a tight-binding fluorescein-scFv complex.
Protein Sci., 14, 2005
|
|
5LY8
 
 | | Structure of the CBM2 module of Lactobacillus casei BL23 phage J-1 evolved Dit. | | Descriptor: | MAGNESIUM ION, Tail component | | Authors: | Cambillau, C, Spinelli, S, Dieterle, M.-E, Piuri, M. | | Deposit date: | 2016-09-26 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Evolved distal tail carbohydrate binding modules of Lactobacillus phage J-1: a novel type of anti-receptor widespread among lactic acid bacteria phages.
Mol. Microbiol., 104, 2017
|
|
4V96
 
 | | The structure of a 1.8 MDa viral genome injection device suggests alternative infection mechanisms | | Descriptor: | BPP, ORF46, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V, van Sinderen, D, Cambillau, C. | | Deposit date: | 2012-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V5I
 
 | | Structure of the Phage P2 Baseplate in its Activated Conformation with Ca | | Descriptor: | CALCIUM ION, ORF15, ORF16, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2010-02-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (5.464 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2PVS
 
 | | Structure of human pancreatic lipase related protein 2 mutant N336Q | | Descriptor: | CALCIUM ION, Pancreatic lipase-related protein 2, SULFATE ION | | Authors: | Spinelli, S, Eydoux, C, Carriere, F, Cambillau, C. | | Deposit date: | 2007-05-10 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human pancreatic lipase-related protein 2 with the lid in an open conformation.
Biochemistry, 47, 2008
|
|
4R96
 
 | | Structure of a Llama Glama Fab 48A2 against human cMet | | Descriptor: | Llama glama Fab 48A2 against human cMet H chain, Llama glama Fab 48A2 against human cMet L chain | | Authors: | Klarenbeek, A, El Mazouari, K, Desmyter, A, Blanchetot, C, Hultberg, A, Roovers, R.C, Cambillau, C, Spinelli, S, Del-Favero, J, Verrips, T, de Haard, H, Achour, I. | | Deposit date: | 2014-09-03 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Camelid Ig V genes reveal significant human homology not seen in therapeutic target genes, providing for a powerful therapeutic antibody platform.
MAbs, 7, 2015
|
|
1LCO
 
 | |
1LPA
 
 | | INTERFACIAL ACTIVATION OF THE LIPASE-PROCOLIPASE COMPLEX BY MIXED MICELLES REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | CALCIUM ION, COLIPASE, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Van Tilbeurgh, H, Egloff, M.-P, Cambillau, C. | | Deposit date: | 1994-08-19 | | Release date: | 1994-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Interfacial activation of the lipase-procolipase complex by mixed micelles revealed by X-ray crystallography.
Nature, 362, 1993
|
|
1LPB
 
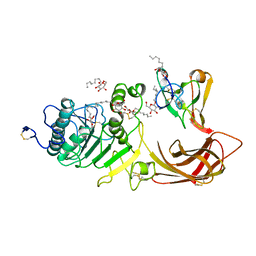 | | THE 2.46 ANGSTROMS RESOLUTION STRUCTURE OF THE PANCREATIC LIPASE COLIPASE COMPLEX INHIBITED BY A C11 ALKYL PHOSPHONATE | | Descriptor: | CALCIUM ION, COLIPASE, LIPASE, ... | | Authors: | Egloff, M.-P, Van Tilbeurgh, H, Cambillau, C. | | Deposit date: | 1994-08-19 | | Release date: | 1994-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The 2.46 A resolution structure of the pancreatic lipase-colipase complex inhibited by a C11 alkyl phosphonate.
Biochemistry, 34, 1995
|
|
4Y7M
 
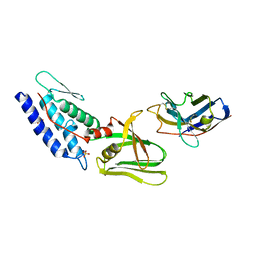 | | T6SS protein TssM C-terminal domain (835-1129) from EAEC | | Descriptor: | Hi113 protein, SULFATE ION, Type VI secretion protein IcmF | | Authors: | Nguyen, V.S, Spinelli, S, Durand, E, Roussel, A, Cambillau, C. | | Deposit date: | 2015-02-15 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biogenesis and structure of a type VI secretion membrane core complex.
Nature, 523, 2015
|
|
4RGA
 
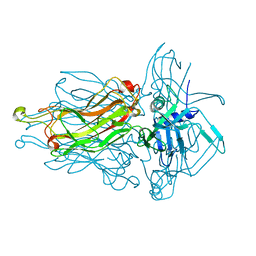 | | Phage 1358 receptor binding protein in complex with the trisaccharide GlcNAc-Galf-GlcOMe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-galactofuranose-(1-6)-methyl alpha-D-glucopyranoside, Phage 1358 receptor binding protein (ORF20) | | Authors: | Spinelli, S, Mccabe, O, Farenc, C, Tremblay, D, Blangy, S, Oscarson, S, Moineau, S, Cambillau, C. | | Deposit date: | 2014-09-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The targeted recognition of Lactococcus lactis phages to their polysaccharide receptors.
Mol.Microbiol., 96, 2015
|
|
6ZIG
 
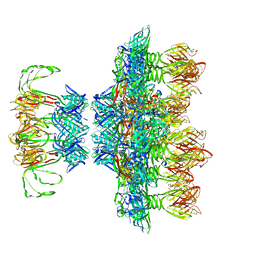 | |
6ZIH
 
 | |
6ZJJ
 
 | |
1XZA
 
 | |
1XZB
 
 | |
1LGB
 
 | |
1LOB
 
 | |
1LOA
 
 | |
6N38
 
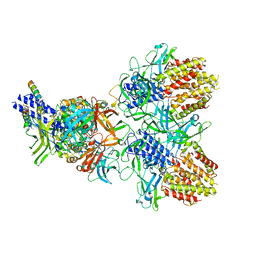 | | Structure of the type VI secretion system TssK-TssF-TssG baseplate subcomplex revealed by cryo-electron microscopy - full map sharpened | | Descriptor: | Putative type VI secretion protein, Unassigned protein | | Authors: | Park, Y.J, Lacourse, K.D, Cambillau, C, Seattle Structural Genomics Center for Infectious Disease (SSGCID), DiMaio, F, Mougous, J.D, Veesler, D. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the type VI secretion system TssK-TssF-TssG baseplate subcomplex revealed by cryo-electron microscopy.
Nat Commun, 9, 2018
|
|
1AGY
 
 | |
1XZJ
 
 | |
1XZC
 
 | |
1XZE
 
 | |
