2IVE
 
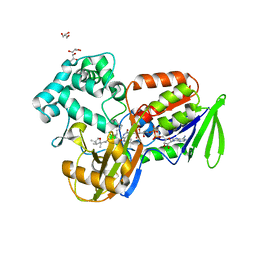 | | Structure of protoporphyrinogen oxidase from Myxococcus xanthus | | Descriptor: | (3S)-3-[(2S,3S,4R)-3,4-DIMETHYLTETRAHYDROFURAN-2-YL]BUTYL LAURATE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Corradi, H.R, Corrigall, A.V, Boix, E, Mohan, C.G, Sturrock, E.D, Meissner, P.N, Acharya, K.R. | | Deposit date: | 2006-06-13 | | Release date: | 2006-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Protoporphyrinogen Oxidase from Myxococcus Xanthus and its Complex with the Inhibitor Acifluorfen.
J.Biol.Chem., 281, 2006
|
|
2IVD
 
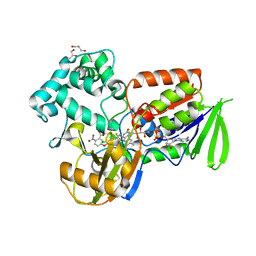 | | Structure of protoporphyrinogen oxidase from Myxococcus xanthus with acifluorfen | | Descriptor: | (3S)-3-[(2S,3S,4R)-3,4-DIMETHYLTETRAHYDROFURAN-2-YL]BUTYL LAURATE, 5-[2-CHLORO-4-(TRIFLUOROMETHYL)PHENOXY]-2-NITROBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Corradi, H.R, Corrigall, A.V, Boix, E, Mohan, C.G, Sturrock, E.D, Meissner, P.N, Acharya, K.R. | | Deposit date: | 2006-06-12 | | Release date: | 2006-10-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Protoporphyrinogen Oxidase from Myxococcus Xanthus and its Complex with the Inhibitor Acifluorfen.
J.Biol.Chem., 281, 2006
|
|
2KB5
 
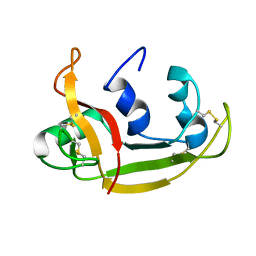 | | Solution NMR Structure of Eosinophil Cationic Protein/RNase 3 | | Descriptor: | Eosinophil cationic protein | | Authors: | Rico, M, Bruix, M, Laurents, D.V, Santoro, J, Jimenez, M, Boix, E, Moussaoui, M, Nogues, M. | | Deposit date: | 2008-11-20 | | Release date: | 2009-06-23 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The (1)H, (13)C, (15)N resonance assignment, solution structure, and residue level stability of eosinophil cationic protein/RNase 3 determined by NMR spectroscopy
Biopolymers, 91, 2009
|
|
1HI5
 
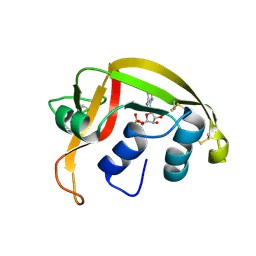 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI3
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine 2'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI4
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosien-3'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI2
 
 | | Eosinophil-derived Neurotoxin (EDN) - Sulphate Complex | | Descriptor: | EOSINOPHIL-DERIVED NEUROTOXIN, SULFATE ION | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1VZT
 
 | | ROLES OF INDIVIDUAL RESIDUES OF ALPHA-1,3 GALACTOSYLTRANSFERASES IN SUBSTRATE BINDING AND CATALYSIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, X, Acharya, K.R, Brew, K. | | Deposit date: | 2004-05-26 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of Individual Enzyme-Substrate Interactions by Alpha-1,3-Galactosyltransferase in Catalysis and Specificity.
Biochemistry, 42, 2003
|
|
1VZU
 
 | | Roles of active site tryptophans in substrate binding and catalysis by ALPHA-1,3 GALACTOSYLTRANSFERASE | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Deshpande, A, Xie, Z, Natesh, R, Acharya, K.R, Brew, K. | | Deposit date: | 2004-05-27 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Roles of active site tryptophans in substrate binding and catalysis by alpha-1,3 galactosyltransferase.
Glycobiology, 14, 2004
|
|
1VZX
 
 | | Roles of active site tryptophans in substrate binding and catalysis by ALPHA-1,3 GALACTOSYLTRANSFERASE | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Deshpande, A, Xie, Z, Natesh, R, Acharya, K.R, Brew, K. | | Deposit date: | 2004-05-28 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Roles of active site tryptophans in substrate binding and catalysis by alpha-1,3 galactosyltransferase.
Glycobiology, 14, 2004
|
|
1O7Q
 
 | | Roles of Individual Residues of Alpha-1,3 Galactosyltransferases in Substrate Binding and Catalysis | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, Z, Acharya, K.R, Brew, K. | | Deposit date: | 2002-11-12 | | Release date: | 2003-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Roles of Individual Enzyme-Substrate Interactions by Alpha-1,3-Galactosyltransferase in Catalysis and Specificity.
Biochemistry, 42, 2003
|
|
1O7O
 
 | | Roles of Individual Residues of Alpha-1,3 Galactosyltransferases in Substrate Binding and Catalysis | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, Z, Acharya, K.R, Brew, K. | | Deposit date: | 2002-11-11 | | Release date: | 2003-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Roles of individual enzyme-substrate interactions by alpha-1,3-galactosyltransferase in catalysis and specificity.
Biochemistry, 42, 2003
|
|
5E13
 
 | | Crystal structure of Eosinophil-derived neurotoxin in complex with the triazole double-headed ribonucleoside 11c | | Descriptor: | 3'-{4-[(4-amino-2-oxopyrimidin-1(2H)-yl)methyl]-1H-1,2,3-triazol-1-yl}-3'-deoxyadenosine, Non-secretory ribonuclease | | Authors: | Chatzileontiadou, D.S.M, Stravodimos, G.A, Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Triazole double-headed ribonucleosides as inhibitors of eosinophil derived neurotoxin.
Bioorg.Chem., 63, 2015
|
|
3GLJ
 
 | | A polymorph of carboxypeptidase B zymogen structure | | Descriptor: | Carboxypeptidase B, GLYCEROL, ZINC ION | | Authors: | Fernandez, D. | | Deposit date: | 2009-03-12 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Analysis of a new crystal form of procarboxypeptidase B: further insights into the catalytic mechanism
Biopolymers, 2009
|
|
3HUV
 
 | |
1GQV
 
 | | Atomic Resolution (0.98A) Structure of Eosinophil-Derived Neurotoxin | | Descriptor: | ACETATE ION, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Swaminathan, G.J, Holloway, D.E, Veluraja, K, Acharya, K.R. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution (0.98 A) Structure of Eosinophil-Derived Neurotoxin
Biochemistry, 41, 2002
|
|
