5D3Q
 
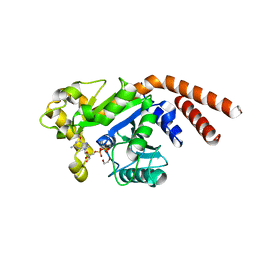 | | Dynamin 1 GTPase-BSE fusion dimer complexed with GDP | | 分子名称: | 1,2-ETHANEDIOL, Dynamin-1,Dynamin-1, GUANOSINE-5'-DIPHOSPHATE | | 著者 | Anand, R, Eschenburg, S, Reubold, T.F. | | 登録日 | 2015-08-06 | | 公開日 | 2015-12-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the GTPase domain and the bundle signalling element of dynamin in the GDP state.
Biochem.Biophys.Res.Commun., 469, 2016
|
|
1J58
 
 | | Crystal Structure of Oxalate Decarboxylase | | 分子名称: | FORMIC ACID, MAGNESIUM ION, MANGANESE (II) ION, ... | | 著者 | Anand, R, Dorrestein, P.C, Kinsland, C, Begley, T.P, Ealick, S.E. | | 登録日 | 2002-02-25 | | 公開日 | 2002-07-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 A resolution.
Biochemistry, 41, 2002
|
|
1L3J
 
 | | Crystal Structure of Oxalate Decarboxylase Formate Complex | | 分子名称: | FORMIC ACID, MAGNESIUM ION, MANGANESE (II) ION, ... | | 著者 | Anand, R, Dorrestein, P.C, Kinsland, C, Begley, T.P, Ealick, S.E. | | 登録日 | 2002-02-27 | | 公開日 | 2002-07-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 A resolution.
Biochemistry, 41, 2002
|
|
1T4A
 
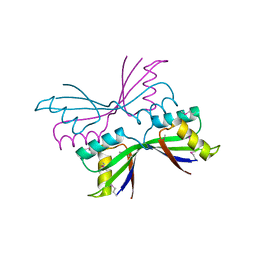 | | Structure of B. Subtilis PurS C2 Crystal Form | | 分子名称: | PurS | | 著者 | Anand, R, Hoskins, A.A, Bennett, E.M, Sintchak, M.D, Stubbe, J, Ealick, S.E. | | 登録日 | 2004-04-28 | | 公開日 | 2004-09-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A model for the Bacillus subtilis formylglycinamide ribonucleotide amidotransferase multiprotein complex.
Biochemistry, 43, 2004
|
|
1TWJ
 
 | |
1S2L
 
 | |
1S2G
 
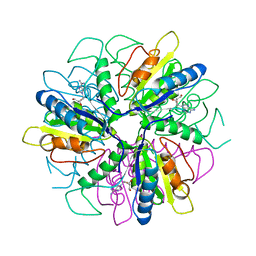 | | Purine 2'deoxyribosyltransferase + 2'-deoxyadenosine | | 分子名称: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, purine trans deoxyribosylase | | 著者 | Anand, R, Kaminski, P.A, Ealick, S.E. | | 登録日 | 2004-01-08 | | 公開日 | 2004-03-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structures of purine 2'-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 A resolution.
Biochemistry, 43, 2004
|
|
1S2I
 
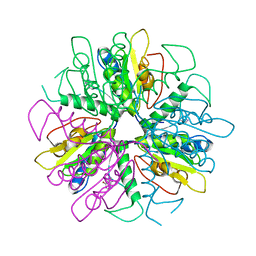 | | Purine 2'deoxyribosyltransferase + bromopurine | | 分子名称: | 6-BROMO-7H-PURINE, purine trans deoxyribosylase | | 著者 | Anand, R, Kaminski, P.A, Ealick, S.E. | | 登録日 | 2004-01-08 | | 公開日 | 2004-03-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structures of purine 2'-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 A resolution.
Biochemistry, 43, 2004
|
|
1S2D
 
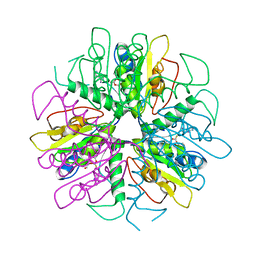 | | Purine 2'-Deoxyribosyl complex with arabinoside: Ribosylated Intermediate (AraA) | | 分子名称: | 2-deoxy-2-fluoro-alpha-D-arabinofuranose, ADENINE, Nucleoside 2-deoxyribosyltransferase | | 著者 | Anand, R, Kaminski, P.A, Ealick, S.E. | | 登録日 | 2004-01-08 | | 公開日 | 2004-03-30 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structures of purine 2'-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 A resolution.
Biochemistry, 43, 2004
|
|
1S3F
 
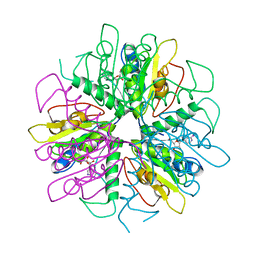 | | Purine 2'-deoxyribosyltransferase + selenoinosine | | 分子名称: | 9-(3,4-DIHYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-2-YL)-1,9-DIHYDRO-PURINE-6-THIONE, purine trans deoxyribosylase | | 著者 | Anand, R, Kaminski, P.A, Ealick, S.E. | | 登録日 | 2004-01-13 | | 公開日 | 2004-03-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structures of purine 2'-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 A resolution.
Biochemistry, 43, 2004
|
|
4HRW
 
 | | Identification of function and Mechanistic insights of Guanine deaminase from Nitrosomonas europaea | | 分子名称: | 5-AMINO-1H-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-7-OL, Cytidine and deoxycytidylate deaminase zinc-binding region, ZINC ION | | 著者 | Anand, R, Bitra, A, Bhukya, H, Tanwar, A.S. | | 登録日 | 2012-10-29 | | 公開日 | 2013-06-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Identification of function and mechanistic insights of guanine deaminase from Nitrosomonas europaea: role of the C-terminal loop in catalysis
Biochemistry, 52, 2013
|
|
4HRQ
 
 | | Identification of Function and Mechanistic Insights of Guanine Deaminase from Nitrosomonas europaea: Role of the C-terminal Loop in Catalysis | | 分子名称: | 5-AMINO-1H-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-7-OL, Cytidine and deoxycytidylate deaminase zinc-binding region, ZINC ION | | 著者 | Anand, R, Bitra, A, Bhukya, H, Tanwar, A.S. | | 登録日 | 2012-10-28 | | 公開日 | 2013-06-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Identification of function and mechanistic insights of guanine deaminase from Nitrosomonas europaea: role of the C-terminal loop in catalysis
Biochemistry, 52, 2013
|
|
3UMM
 
 | | Formylglycinamide ribonucleotide amidotransferase from Salmonella typhimurium: Role of the ATP complexation and glutaminase domain in catalytic coupling | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Anand, R, Morar, M, Tanwar, A.S, Panjikar, S. | | 登録日 | 2011-11-14 | | 公開日 | 2012-06-06 | | 最終更新日 | 2013-09-11 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Formylglycinamide ribonucleotide amidotransferase from Salmonella typhimurium: role of ATP complexation and the glutaminase domain in catalytic coupling
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UJN
 
 | | Formyl Glycinamide Ribonucleotide Amidotransferase from Salmonella Typhimurium : Role of the ATP complexation and glutaminase domain in catalytic coupling | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase, ... | | 著者 | Anand, R, Morar, M, Tanwar, A.S, Panjikar, S. | | 登録日 | 2011-11-08 | | 公開日 | 2012-06-06 | | 最終更新日 | 2013-09-11 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Formylglycinamide ribonucleotide amidotransferase from Salmonella typhimurium: role of ATP complexation and the glutaminase domain in catalytic coupling
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1T3T
 
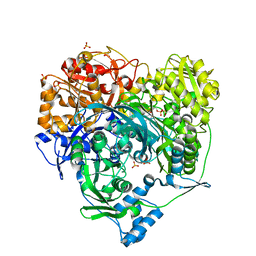 | | Structure of Formylglycinamide synthetase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase, ... | | 著者 | Ealick, S.E, Anand, R, Hoskin, A.A, Stubbe, J. | | 登録日 | 2004-04-27 | | 公開日 | 2004-09-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Domain Organization of Salmonella typhimurium Formylglycinamide Ribonucleotide Amidotransferase Revealed by X-ray crystallography
Biochemistry, 43, 2004
|
|
4LCN
 
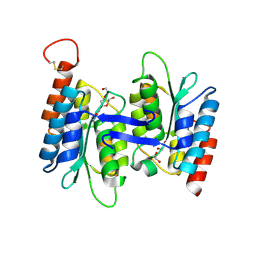 | | Crytsal structure of NE0047 in complex with 2'-DEOXY-GUANOSINE | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXY-GUANOSINE, Cytidine and deoxycytidylate deaminase zinc-binding region, ... | | 著者 | Anand, R, Bitra, A, Biswas, A. | | 登録日 | 2013-06-22 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of the substrate specificity of cytidine deaminase superfamily Guanine deaminase
Biochemistry, 52, 2013
|
|
5XKQ
 
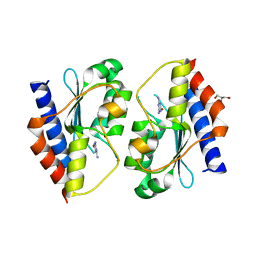 | | Crystal structure of Msmeg3575 in complex with ammeline | | 分子名称: | 4,6-diamino-1,3,5-triazin-2-ol, ACETATE ION, CMP/dCMP deaminase, ... | | 著者 | Gaded, V.M, Anand, R. | | 登録日 | 2017-05-09 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Selective Deamination of Mutagens by a Mycobacterial Enzyme
J. Am. Chem. Soc., 139, 2017
|
|
4R7G
 
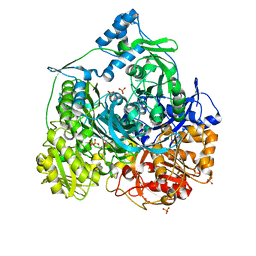 | | Determination of the formylglycinamide ribonucleotide amidotransferase ammonia pathway by combining 3D-RISM theory with experiment | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase, ... | | 著者 | Tanwar, A.S, Sindhikara, D.J, Hirata, F, Anand, R. | | 登録日 | 2014-08-27 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Determination of the formylglycinamide ribonucleotide amidotransferase ammonia pathway by combining 3D-RISM theory with experiment.
Acs Chem.Biol., 10, 2015
|
|
5H58
 
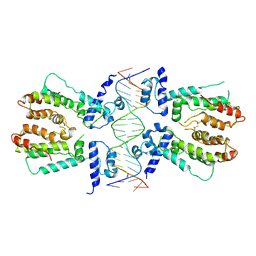 | | Structural and dynamics studies of the TetR family protein, CprB from Streptomyces coelicolor in complex with its biological operator sequence | | 分子名称: | CprB, DNA (5'-D(*AP*GP*GP*C*AP*GP*GP*CP*GP*GP*CP*AP*CP*GP*GP*TP*CP*TP*GP*TP*TP*GP*AP*GP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*A*CP*TP*CP*AP*AP*CP*AP*GP*AP*CP*CP*GP*TP*GP*CP*CP*GP*CP*CP*TP*GP*CP*CP*T)-3') | | 著者 | Bhukya, H, Jana, A.K, Sengupta, N, Anand, R. | | 登録日 | 2016-11-04 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-31 | | 実験手法 | X-RAY DIFFRACTION (3.991 Å) | | 主引用文献 | Structural and dynamics studies of the TetR family protein, CprB from Streptomyces coelicolor in complex with its biological operator sequence
J. Struct. Biol., 198, 2017
|
|
5KBI
 
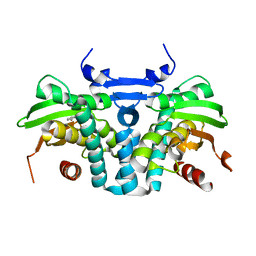 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH CATACHOL | | 分子名称: | CATECHOL, MopR, ZINC ION | | 著者 | Ray, S, Anand, R, Panjikar, S. | | 登録日 | 2016-06-03 | | 公開日 | 2016-07-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
5A3F
 
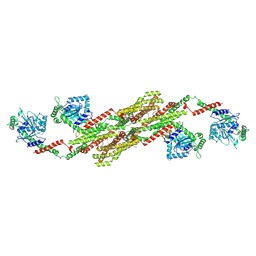 | | Crystal structure of the dynamin tetramer | | 分子名称: | DYNAMIN 3 | | 著者 | Reubold, T.F, Faelber, K, Plattner, N, Posor, Y, Branz, K, Curth, U, Schlegel, J, Anand, R, Manstein, D.J, Noe, F, Haucke, V, Daumke, O, Eschenburg, S. | | 登録日 | 2015-05-29 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Crystal Structure of the Dynamin Tetramer
Nature, 525, 2015
|
|
5KBG
 
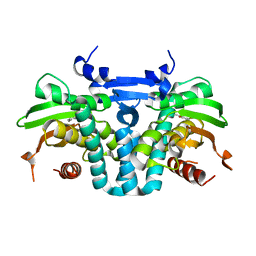 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH OCRESOL | | 分子名称: | MopR, ZINC ION, o-cresol | | 著者 | Ray, S, Anand, R, Panjikar, S. | | 登録日 | 2016-06-03 | | 公開日 | 2016-07-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
5KBE
 
 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH PHENOL | | 分子名称: | MopR, PHENOL, ZINC ION | | 著者 | Ray, S, Anand, R, Panjikar, S. | | 登録日 | 2016-06-03 | | 公開日 | 2016-07-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
5KBH
 
 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH 3-CHLORO-PHENOL | | 分子名称: | 3-CHLOROPHENOL, MopR, ZINC ION | | 著者 | Ray, S, Anand, R, Panjikar, S. | | 登録日 | 2016-06-03 | | 公開日 | 2016-07-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
3CO7
 
 | | Crystal Structure of FoxO1 DBD Bound to DBE2 DNA | | 分子名称: | DNA (5'-D(*DCP*DAP*DAP*DAP*DAP*DTP*DGP*DTP*DAP*DAP*DAP*DCP*DAP*DAP*DGP*DA)-3'), DNA (5'-D(*DTP*DCP*DTP*DTP*DGP*DTP*DTP*DTP*DAP*DCP*DAP*DTP*DTP*DTP*DTP*DG)-3'), Forkhead box protein O1 | | 著者 | Brent, M.M, Anand, R, Marmorstein, R. | | 登録日 | 2008-03-27 | | 公開日 | 2008-09-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Structural Basis for DNA Recognition by FoxO1 and Its Regulation by Posttranslational Modification.
Structure, 16, 2008
|
|
