4HD4
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium V218F mutant | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influencing the monophenolase/diphenolase activity ratio in tyrosinase.
Biochim.Biophys.Acta, 1834, 2013
|
|
4HD7
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium V218G mutant soaked in CuSO4 | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Influencing the monophenolase/diphenolase activity ratio in tyrosinase.
Biochim.Biophys.Acta, 1834, 2013
|
|
5X5I
 
 | | The X-ray crystal structure of a TetR family transcription regulator RcdA involved in the regulation of biofilm formation in Escherichia coli | | Descriptor: | HTH-type transcriptional regulator RcdA | | Authors: | Sugino, H, Usui, M, Shimada, T, Nakano, M, Ogasawara, H, Ishihama, A, Hirata, A. | | Deposit date: | 2017-02-16 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | A structural sketch of RcdA, a transcription factor controlling the master regulator of biofilm formation.
FEBS Lett., 591, 2017
|
|
5GTC
 
 | | Crystal structure of complex between DMAP-SH conjugated with a Kaposi's sarcoma herpesvirus LANA peptide (5-15) and nucleosome core particle | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Kato, D, Suto, H, Kurumizaka, H, Kawashima, S.A, Yamatsugu, K, Kanai, M. | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic Posttranslational Modifications: Chemical Catalyst-Driven Regioselective Histone Acylation of Native Chromatin.
J. Am. Chem. Soc., 139, 2017
|
|
2QZR
 
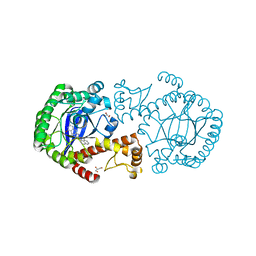 | | tRNA-Guanine Transglycosylase(TGT) in Complex with 6-amino-2-[(1-naphthylmethyl)amino]-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-[(1-naphthylmethyl)amino]-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Hoertner, S.R, Ritschel, T, Stengl, B, Kramer, C, Schweizer, W.B, Wagner, B, Kansy, M, Klebe, G, Diederich, F. | | Deposit date: | 2007-08-17 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Potent Inhibitors of tRNA-Guanine Transglycosylase, an Enzyme Linked to the Pathogenicity of the Shigella Bacterium: Charge-Assisted Hydrogen Bonding.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
3AZ4
 
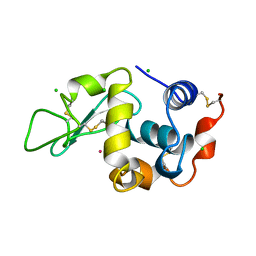 | | Crystal structure of Co/O-HEWL | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Lysozyme C | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
3AZ5
 
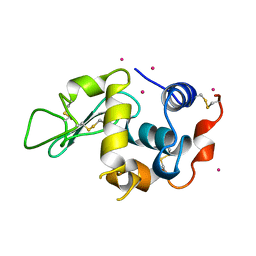 | | Crystal structure of Pt/O-HEWL | | Descriptor: | Lysozyme C, PLATINUM (II) ION | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
3AZ6
 
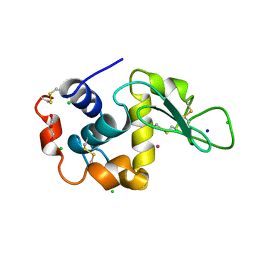 | | Crystal structure of Co/T-HEWL | | Descriptor: | CHLORIDE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
3AZ7
 
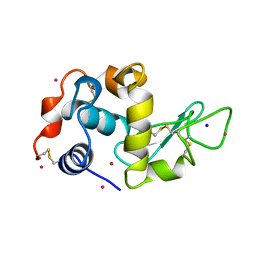 | | Crystal structure of Pt/T-HEWL | | Descriptor: | Lysozyme C, PLATINUM (II) ION, SODIUM ION | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
2E6X
 
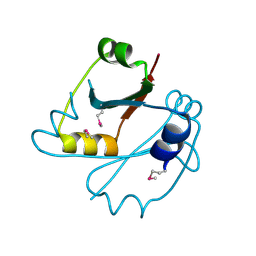 | | X-ray structure of TT1592 from Thermus thermophilus HB8 | | Descriptor: | Hypothetical protein TTHA1281 | | Authors: | Yamada, M, Nakagawa, N, Kanagawa, M, Kuramitsu, S, Kamitori, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-05 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of TT1592 from Thermus thermophilus HB8
To be Published
|
|
2IP4
 
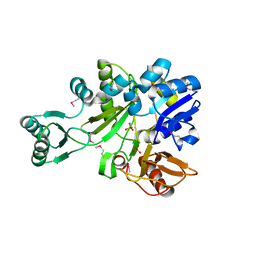 | | Crystal Structure of Glycinamide Ribonucleotide Synthetase from Thermus thermophilus HB8 | | Descriptor: | Phosphoribosylamine--glycine ligase, SULFATE ION | | Authors: | Sampei, G, Baba, S, Kanagawa, M, Yanai, H, Ishii, T, Kawai, H, Fukai, Y, Ebihara, A, Nakagawa, N, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-11 | | Release date: | 2007-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
2Y6L
 
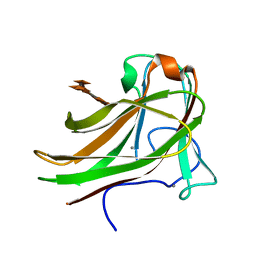 | | Xylopentaose binding X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
3W7B
 
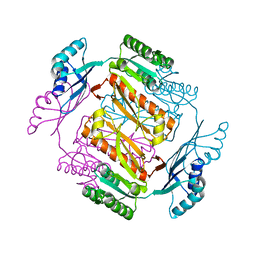 | | Crystal structure of formyltetrahydrofolate deformylase from Thermus thermophilus HB8 | | Descriptor: | Formyltetrahydrofolate deformylase | | Authors: | Sampei, G, Yanagida, Y, Ogata, N, Kusano, M, Terao, K, Kawai, H, Fukai, Y, Kanagawa, M, Inoue, Y, Baba, S, Kawai, G. | | Deposit date: | 2013-02-28 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures and reaction mechanisms of the two related enzymes, PurN and PurU
J.Biochem., 154, 2013
|
|
2Y6J
 
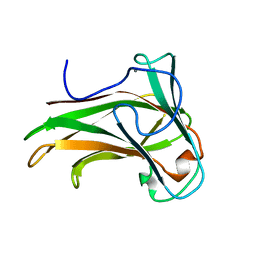 | | X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
1V8B
 
 | | Crystal structure of a hydrolase | | Descriptor: | ADENOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, adenosylhomocysteinase | | Authors: | Tanaka, N, Nakanishi, M, Kusakabe, Y, Shiraiwa, K, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2004-01-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of S-Adenosyl-l-Homocysteine Hydrolase from the Human Malaria Parasite Plasmodium falciparum
J.Mol.Biol., 343, 2004
|
|
2Y6H
 
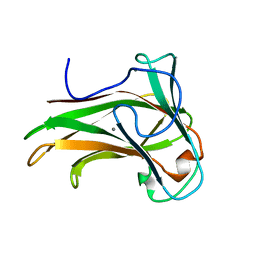 | | X-2 L110F CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-21 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
3ZSM
 
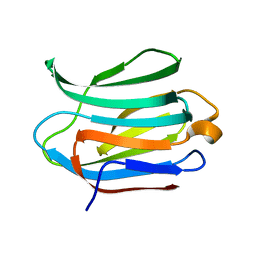 | | Crystal structure of Apo Human Galectin-3 CRD at 1.25 angstrom resolution, at room temperature | | Descriptor: | GALECTIN-3 | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
2Y6G
 
 | | Cellopentaose binding mutated (X-2 L110F) CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-21 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
7E5P
 
 | | Aptamer enhancing peroxidase activity of myoglobin | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*GP*GP*GP*TP*TP*GP*GP*GP*AP*GP*GP*G)-3') | | Authors: | Tsukakoshi, K, Matsugami, A, Khunathai, K, Kanazashi, M, Yamagishi, Y, Nakama, K, Oshikawa, D, Hayashi, F, Kuno, H, Ikebukuro, K. | | Deposit date: | 2021-02-19 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-quadruplex-forming aptamer enhances the peroxidase activity of myoglobin against luminol.
Nucleic Acids Res., 49, 2021
|
|
3ZSK
 
 | | Crystal structure of Human Galectin-3 CRD with glycerol bound at 0.90 angstrom resolution | | Descriptor: | GALECTIN-3, GLYCEROL | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
3ZSL
 
 | | Crystal structure of Apo Human Galectin-3 CRD at 1.08 angstrom resolution, at cryogenic temperature | | Descriptor: | GALECTIN-3 | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
4A6R
 
 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in the apo form, crystallised from polyacrylic acid | | Descriptor: | OMEGA TRANSAMINASE, POLYACRYLIC ACID | | Authors: | Logan, D.T, Hakansson, M, Yengo, K, Svedendahl Humble, M, Engelmark Cassimjee, K, Walse, B, Abedi, V, Federsel, H.-J, Berglund, P. | | Deposit date: | 2011-11-08 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Crystal Structures of the Chromobacterium Violaceum Omega-Transaminase Reveal Major Structural Rearrangements Upon Binding of Coenzyme Plp.
FEBS J., 279, 2012
|
|
4A72
 
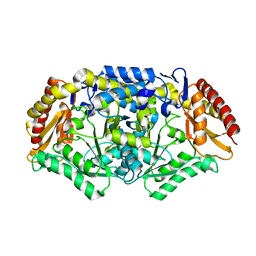 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in a mixture of apo and PLP-bound states | | Descriptor: | OMEGA TRANSAMINASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Logan, D.T, Hakansson, M, Yengo, K, Svedendahl Humble, M, Engelmark Cassimjee, K, Walse, B, Abedi, V, Federsel, H.-J, Berglund, P. | | Deposit date: | 2011-11-10 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Chromobacterium Violaceum Omega-Transaminase Reveal Major Structural Rearrangements Upon Binding of Coenzyme Plp.
FEBS J., 279, 2012
|
|
2Y6K
 
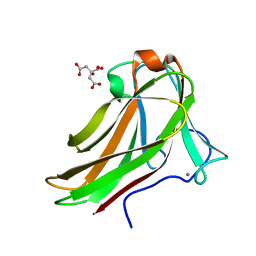 | | Xylotetraose bound to X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, CITRIC ACID, XYLANASE, ... | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
4A6U
 
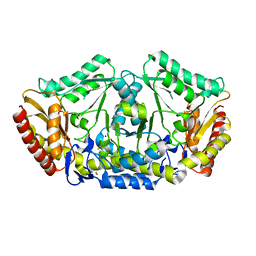 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in the apo form, crystallised from PEG 3350 | | Descriptor: | OMEGA TRANSAMINASE, SODIUM ION, THIOCYANATE ION | | Authors: | Logan, D.T, Hakansson, M, Yengo, K, Svedendahl Humble, M, Engelmark Cassimjee, K, Walse, B, Abedi, V, Federsel, H.-J, Berglund, P. | | Deposit date: | 2011-11-08 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.687 Å) | | Cite: | Crystal Structures of the Chromobacterium Violaceum Omega-Transaminase Reveal Major Structural Rearrangements Upon Binding of Coenzyme Plp.
FEBS J., 279, 2012
|
|
