1W62
 
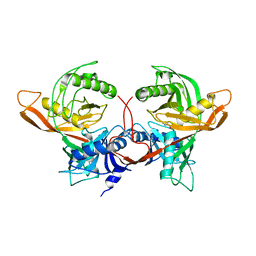 | |
8P5T
 
 | | Single particle cryo-EM structure of the homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, MAGNESIUM ION, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
4DEC
 
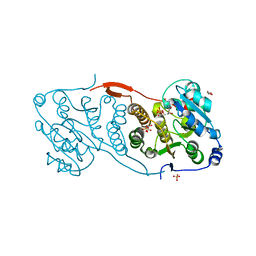 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+, uridine-diphosphate (UDP) and phosphoglyceric acid (PGA) | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, GLUCOSYL-3-PHOSPHOGLYCERATE SYNTHASE (GpgS), GLYCEROL, ... | | Authors: | Albesa-Jove, D, Urresti, S, van der Woerd, M, Guerin, M.E. | | Deposit date: | 2012-01-20 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Mechanistic insights into the retaining glucosyl-3-phosphoglycerate synthase from mycobacteria.
J.Biol.Chem., 287, 2012
|
|
6ZZJ
 
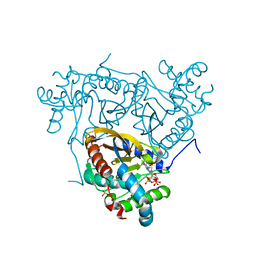 | | Crystal structure of the catalytic domain of Corynebacterium glutamicum acetyltransferase AceF (E2p) in complex with oxidized CoA. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, OXIDIZED COENZYME A | | Authors: | Bruch, E.M, Lexa-Sapart, N, Bellinzoni, M. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Actinobacteria challenge the paradigm: A unique protein architecture for a well-known, central metabolic complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
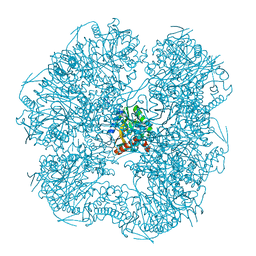
6ZZN
 
 | |
6ZZK
 
 | |
6ZZM
 
 | |
6ZZL
 
 | |
6ZZI
 
 | |
1A4E
 
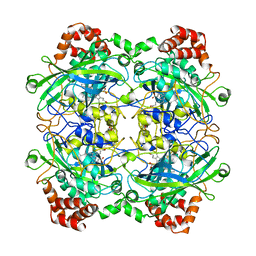 | | CATALASE A FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | AZIDE ION, CATALASE A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mate, M.J. | | Deposit date: | 1998-01-29 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of catalase-A from Saccharomyces cerevisiae.
J.Mol.Biol., 286, 1999
|
|
2C8T
 
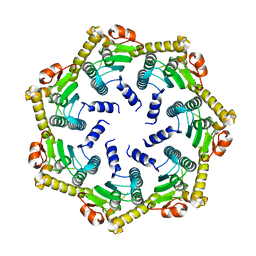 | | The 3.0 A Resolution Structure of Caseinolytic Clp Protease 1 from Mycobacterium tuberculosis | | Descriptor: | ATP-DEPENDENT CLP PROTEASE PROTEOLYTIC SUBUNIT 1 | | Authors: | Ingvarsson, H, Hogbom, M, Jones, T.A, Unge, T. | | Deposit date: | 2005-12-07 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights Into the Inter-Ring Plasticity of Caseinolytic Proteases from the X-Ray Structure of Mycobacterium Tuberculosis Clpp1.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2FDQ
 
 | |
2IHL
 
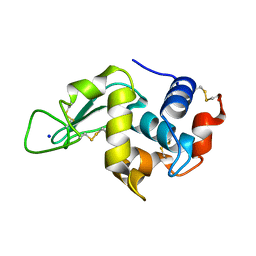 | | LYSOZYME (E.C.3.2.1.17) (JAPANESE QUAIL) | | Descriptor: | JAPANESE QUAIL EGG WHITE LYSOZYME, SODIUM ION | | Authors: | Houdusse, A, Bentley, G.A, Poljak, R.J, Souchon, H, Zhang, Z. | | Deposit date: | 1993-06-29 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Three-dimensional structure of a heteroclitic antigen-antibody cross-reaction complex.
Proc.Natl.Acad.Sci.Usa, 90, 1993
|
|
6C6I
 
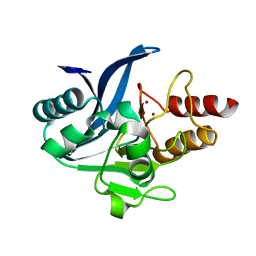 | | Crystal structure of a chimeric NDM-1 metallo-beta-lactamase harboring the IMP-1 L3 loop | | Descriptor: | Metallo-beta-lactamase type 2 chimera, ZINC ION | | Authors: | Otero, L, Giannini, E, Klinke, S, Palacios, A, Mojica, M, Bonomo, R, Llarrull, L, Vila, A. | | Deposit date: | 2018-01-18 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Reaction Mechanism of Metallo-beta-Lactamases Is Tuned by the Conformation of an Active-Site Mobile Loop.
Antimicrob. Agents Chemother., 63, 2019
|
|
3EHG
 
 | |
1AFS
 
 | | RECOMBINANT RAT LIVER 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE (3-ALPHA-HSD) COMPLEXED WITH NADP AND TESTOSTERONE | | Descriptor: | 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TESTOSTERONE | | Authors: | Bennett, M.J, Albert, R.H, Jez, J.M, Ma, H, Penning, T.M, Lewis, M. | | Deposit date: | 1997-03-13 | | Release date: | 1997-10-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Steroid recognition and regulation of hormone action: crystal structure of testosterone and NADP+ bound to 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase.
Structure, 5, 1997
|
|
4DDZ
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis | | Descriptor: | GLUCOSYL-3-PHOSPHOGLYCERATE SYNTHASE (GpgS), GLYCEROL | | Authors: | Albesa-Jove, D, Urresti, S, Gest, P.M, van der Woerd, M, Jackson, M, Guerin, M.E. | | Deposit date: | 2012-01-19 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanistic insights into the retaining glucosyl-3-phosphoglycerate synthase from mycobacteria.
J.Biol.Chem., 287, 2012
|
|
2WE4
 
 | | Carbamate kinase from Enterococcus faecalis bound to a sulfate ion and two water molecules, which mimic the substrate carbamyl phosphate | | Descriptor: | CARBAMATE KINASE 1, SULFATE ION | | Authors: | Ramon-Maiques, S, Marina, A, Gil-Ortiz, F, Rubio, V. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Substrate Binding and Catalysis in Carbamate Kinase Ascertained by Crystallographic and Site-Directed Mutagenesis Studies. Movements and Significance of a Unique Globular Subdomain of This Key Enzyme for Fermentative ATP Production in Bacteria.
J.Mol.Biol., 397, 2010
|
|
2WE5
 
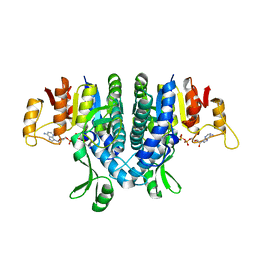 | | Carbamate kinase from Enterococcus faecalis bound to MgADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CARBAMATE KINASE 1, ... | | Authors: | Ramon-Maiques, S, Marina, A, Rubio, V. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Substrate Binding and Catalysis in Carbamate Kinase Ascertained by Crystallographic and Site- Directed Mutagenesis Studies. Movements and Significance of a Unique Globular Subdomain of This Key Enzyme for Fermentative ATP Production in Bacteria.
J.Mol.Biol., 397, 2010
|
|
2GV0
 
 | |
4DE7
 
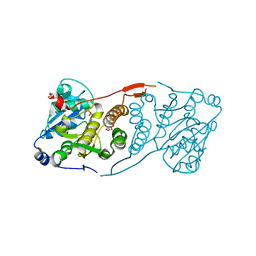 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mg2+ and uridine-diphosphate (UDP) | | Descriptor: | GLUCOSYL-3-PHOSPHOGLYCERATE SYNTHASE (GpgS), GLYCEROL, URIDINE-5'-DIPHOSPHATE | | Authors: | Albesa-Jove, D, Urresti, S, van der Woerd, M, Guerin, M.E. | | Deposit date: | 2012-01-20 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanistic insights into the retaining glucosyl-3-phosphoglycerate synthase from mycobacteria.
J.Biol.Chem., 287, 2012
|
|
1LWI
 
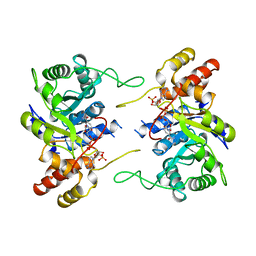 | | 3-ALPHA-HYDROXYSTEROID/DIHYDRODIOL DEHYDROGENASE FROM RATTUS NORVEGICUS | | Descriptor: | 3-ALPHA-HYDROXYSTEROID/DIHYDRODIOL DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bennett, M.J, Schlegel, B.P, Jez, J.M, Penning, T.M, Lewis, M. | | Deposit date: | 1996-02-24 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase complexed with NADP+.
Biochemistry, 35, 1996
|
|
