1L61
 
 | |
1L39
 
 | |
1L62
 
 | |
1L73
 
 | |
1L66
 
 | |
1L46
 
 | |
1L65
 
 | |
1L72
 
 | |
1L57
 
 | |
1L42
 
 | |
1L59
 
 | |
1L70
 
 | |
1L75
 
 | |
1YSA
 
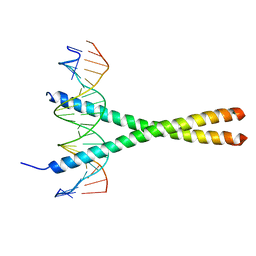 | | THE GCN4 BASIC REGION LEUCINE ZIPPER BINDS DNA AS A DIMER OF UNINTERRUPTED ALPHA HELICES: CRYSTAL STRUCTURE OF THE PROTEIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*CP*TP*GP*GP*AP*TP*GP*AP*GP*TP*CP*AP*TP*A P*GP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*CP*TP*AP*TP*GP*AP*CP*TP*CP*AP*TP*CP*CP*A P*GP*TP*T)-3'), PROTEIN (GCN4) | | Authors: | Ellenberger, T.E, Brandl, C.J, Struhl, K, Harrison, S.C. | | Deposit date: | 1993-08-09 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex.
Cell(Cambridge,Mass.), 71, 1992
|
|
4JL5
 
 | |
4CF7
 
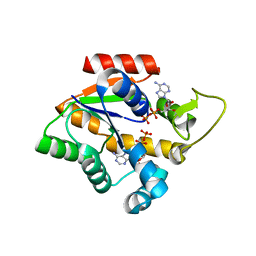 | | Crystal structure of adenylate kinase from Aquifex aeolicus with MgADP bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINASE, ... | | Authors: | Kerns, S.J, Agafonov, R.V, Cho, Y.-J, Pontiggia, F, Otten, R, Pachov, D.V, Kutter, S, Phung, L.A, Murphy, P.N, Thai, V, Hagan, M.F, Kern, D. | | Deposit date: | 2013-11-13 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | The Energy Landscape of Adenylate Kinase During Catalysis.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4JL6
 
 | |
4JLO
 
 | |
4JLA
 
 | |
4JKY
 
 | |
4JLB
 
 | |
4JLP
 
 | |
4JL8
 
 | |
4JLD
 
 | |
1L24
 
 | |
