1VC2
 
 | |
1DI3
 
 | |
1DI5
 
 | |
1VD6
 
 | |
1DI4
 
 | |
3X3U
 
 | |
2ZWB
 
 | | Neutron crystal structure of wild type human lysozyme in D2O | | Descriptor: | Lysozyme C | | Authors: | Chiba-Kamoshida, K, Matsui, T, Chatake, T, Ohhara, T, Ostermann, A, Tanaka, I, Yutani, K, Niimura, N. | | Deposit date: | 2008-12-02 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | Site-specific softening of peptide bonds by localized deuterium observed by neutron crystallography of human lysozyme
To be Published
|
|
3AA9
 
 | | Crystal Structure Analysis of the Mutant CutA1 (E61V) from E. coli | | Descriptor: | Divalent-cation tolerance protein cutA | | Authors: | Matsuura, Y, Tanaka, T, Bagautdinov, B, Kunishima, N, Yutani, K. | | Deposit date: | 2009-11-12 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design
J.Biochem., 148, 2010
|
|
2DZU
 
 | |
2DZS
 
 | |
2DZW
 
 | |
2E09
 
 | |
2DZP
 
 | |
2DZX
 
 | |
3AA8
 
 | | Crystal Structure Analysis of the Mutant CutA1 (S11V/E61V) from E. coli | | Descriptor: | Divalent-cation tolerance protein cutA | | Authors: | Matsuura, Y, Tanaka, T, Bagautdinov, B, Kunishima, N, Yutani, K. | | Deposit date: | 2009-11-12 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design
J.Biochem., 148, 2010
|
|
3AH6
 
 | | Remarkable improvement of the heat stability of CutA1 from E.coli by rational protein designing | | Descriptor: | Divalent-cation tolerance protein cutA | | Authors: | Matsuura, Y, Tanaka, T, Bagautdinov, B, Kunishima, N, Yutani, K. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design
J.Biochem., 148, 2010
|
|
2DZV
 
 | |
2DZT
 
 | |
1WN1
 
 | | Crystal Structure of Dipeptiase from Pyrococcus Horikoshii OT3 | | Descriptor: | COBALT (II) ION, dipeptidase | | Authors: | Jeyakanthan, J, Taka, J, Kitaguchi, Y, Shiro, Y, Yokoyama, S, Yutani, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-26 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Dipeptiase from Pyrococcus Horikoshii OT3
To be Published
|
|
1IOI
 
 | | x-ray crystalline structures of pyrrolidone carboxyl peptidase from a hyperthermophile, pyrococcus furiosus, and its cys-free mutant | | Descriptor: | PYRROLIDONE CARBOXYL PEPTIDASE | | Authors: | Tanaka, H, Chinami, M, Ota, M, Tsukihara, T, Yutani, K. | | Deposit date: | 2001-03-09 | | Release date: | 2001-03-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray crystalline structures of pyrrolidone carboxyl peptidase from a hyperthermophile, Pyrococcus furiosus, and its cys-free mutant.
J.Biochem., 130, 2001
|
|
1CKH
 
 | | T70V MUTANT HUMAN LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME) | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
1CKD
 
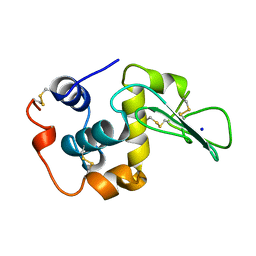 | | T43V MUTANT HUMAN LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
1CJ9
 
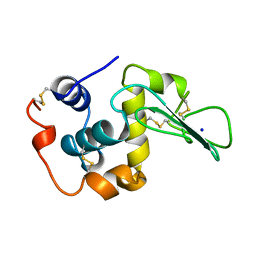 | | T40V MUTANT HUMAN LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
1CKC
 
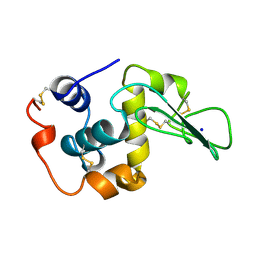 | | T43A MUTANT HUMAN LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
1CKF
 
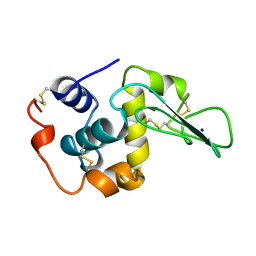 | | T52A MUTANT HUMAN LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
