8CWH
 
 | | 200us Temperature-Jump (Dark2) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CVU
 
 | | 20ns Temperature-Jump (Light) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW8
 
 | | Laser Off Temperature-Jump XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW5
 
 | | 200us Temperature-Jump (Light) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW1
 
 | | 20us Temperature-Jump (Dark1) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWG
 
 | | 200us Temperature-Jump (Dark1) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWC
 
 | | 20ns Temperature-Jump (Light) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW7
 
 | | 200us Temperature-Jump (Dark2) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW6
 
 | | 200us Temperature-Jump (Dark1) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW3
 
 | | 20us Temperature-Jump (Dark2) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWB
 
 | | Laser Off Temperature-Jump XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWE
 
 | | 20ns Temperature-Jump (Dark2) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CVV
 
 | | 20ns Temperature-Jump (Dark1) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWD
 
 | | 20ns Temperature-Jump (Dark1) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CVW
 
 | | 20ns Temperature-Jump (Dark2) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWF
 
 | | 200us Temperature-Jump (Light) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8WZV
 
 | | SFX structure of an Mn-carbonyl complex immobilized in hen egg white lysozyme microcrystals, 1 microsecond after photoexcitation with 40 microJoules laser intensity at RT. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Maity, B, Shoji, M, Luo, F, Nakane, T, Abe, S, Owada, S, Kang, J, Tono, K, Tanaka, R, Thuc, T.T, Kojima, M, Hishikawa, Y, Tanaka, J, Tian, J, Noya, H, Nakasuji, Y, Asanuma, A, Yao, X, Iwata, S, Shigeta, Y, Nango, E, Ueno, T. | | Deposit date: | 2023-11-02 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Real-time observation of a metal complex-driven reaction intermediate using a porous protein crystal and serial femtosecond crystallography.
Nat Commun, 15, 2024
|
|
8WZT
 
 | | SFX structure of an Mn-carbonyl complex immobilized in hen egg white lysozyme microcrystals, 1 microsecond after photoexcitation at RT. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Maity, B, Shoji, M, Luo, F, Nakane, T, Abe, S, Owada, S, Kang, J, Tono, K, Tanaka, R, Thuc, T.T, Kojima, M, Hishikawa, Y, Tanaka, J, Tian, J, Noya, H, Nakasuji, Y, Asanuma, A, Yao, X, Iwata, S, Shigeta, Y, Nango, E, Ueno, T. | | Deposit date: | 2023-11-02 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Real-time observation of a metal complex-driven reaction intermediate using a porous protein crystal and serial femtosecond crystallography.
Nat Commun, 15, 2024
|
|
8WZF
 
 | | Dark state SFX structure of hen egg white lysozyme microcrystals immobilized with a Mn carbonyl complex at RT. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Maity, B, Shoji, M, Luo, F, Nakane, T, Abe, S, Owada, S, Kang, J, Tono, K, Tanaka, R, Thuc, T.T, Kojima, M, Hishikawa, Y, Tanaka, J, Tian, J, Noya, H, Nakasuji, Y, Asanuma, A, Yao, X, Iwata, S, Shigeta, Y, Nango, E, Ueno, T. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Real-time observation of a metal complex-driven reaction intermediate using a porous protein crystal and serial femtosecond crystallography.
Nat Commun, 15, 2024
|
|
8AJZ
 
 | | Serial femtosecond crystallography structure of CO bound ba3- type cytochrome c oxidase at 2 milliseconds after irradiation by a 532 nm laser | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Bath, P, Bosman, R, Dahl, P, Nango, E, Tanaka, R, Zoric, D, Svensson, E, Nakane, T, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
8WZG
 
 | | SFX structure of an Mn-carbonyl complex immobilized in hen egg white lysozyme microcrystals, 10 ns after photoexcitation at RT. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Maity, B, Shoji, M, Luo, F, Nakane, T, Abe, S, Owada, S, Kang, J, Tono, K, Tanaka, R, Thuc, T.T, Kojima, M, Hishikawa, Y, Tanaka, J, Tian, J, Noya, H, Nakasuji, Y, Asanuma, A, Yao, X, Iwata, S, Shigeta, Y, Nango, E, Ueno, T. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Real-time observation of a metal complex-driven reaction intermediate using a porous protein crystal and serial femtosecond crystallography.
Nat Commun, 15, 2024
|
|
8WZR
 
 | | SFX structure of an Mn-carbonyl complex immobilized in hen egg white lysozyme microcrystals, 100 ns after photoexcitation at RT. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Maity, B, Shoji, M, Luo, F, Nakane, T, Abe, S, Owada, S, Kang, J, Tono, K, Tanaka, R, Thuc, T.T, Kojima, M, Hishikawa, Y, Tanaka, J, Tian, J, Noya, H, Nakasuji, Y, Asanuma, A, Yao, X, Iwata, S, Shigeta, Y, Nango, E, Ueno, T. | | Deposit date: | 2023-11-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Real-time observation of a metal complex-driven reaction intermediate using a porous protein crystal and serial femtosecond crystallography.
Nat Commun, 15, 2024
|
|
6GJW
 
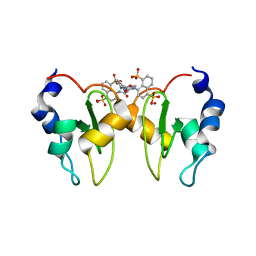 | | Structure of XIAP-BIR1 domain in complex with an NF023 analog | | Descriptor: | 4-[[3-[[3-[(4,8-disulfonatonaphthalen-1-yl)carbamoyl]phenyl]carbamoylamino]phenyl]carbonylamino]naphthalene-1,5-disulfonate, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Sorrentino, L, Cossu, F, Malkoc, B, Zaffaroni, M, Milani, M, Mastrangelo, E. | | Deposit date: | 2018-05-17 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Activity Relationship of NF023 Derivatives Binding to XIAP-BIR1.
Chemistryopen, 8, 2019
|
|
8A6E
 
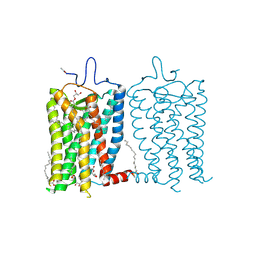 | | 100 picosecond light activated crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SACLA) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C.J, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P.J.M, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C.M, Diethelm, A.D, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-06-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
8A6D
 
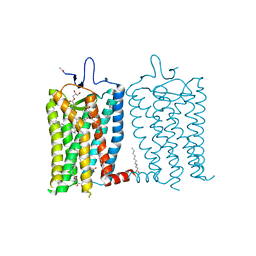 | | 10 picosecond light activated crystal structure of bovine rhodopsin in Lipidic Cubic Phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C.J, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P.J.M, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C.M, Diethelm, A.D, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-06-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
