7ZSS
 
 | | cryo-EM structure of D614 spike in complex with de novo designed binder | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-08 | | Release date: | 2023-03-01 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZRV
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, full map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, ... | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
8OZI
 
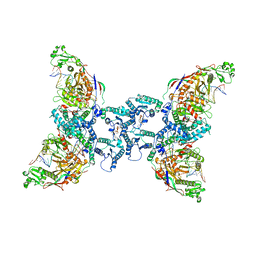 | | cryoEM structure of SPARTA complex pre-NAD cleavage | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Piwi domain-containing protein, ... | | Authors: | Babatunde, E, Dong, C.N, Xu, H.L, Henning, S. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
8OZE
 
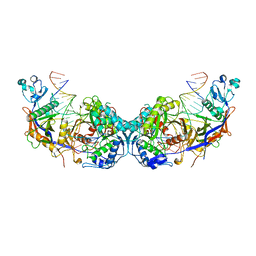 | | cryoEM structure of SPARTA complex dimer high resolution | | Descriptor: | DNA (5'-D(P*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*C)-3'), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*G)-3'), ... | | Authors: | Babatunde, E, Dong, C.N, Xu, H.L, Henning, S. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
8OZG
 
 | | cryoEM structure of SPARTA complex Tetramer Post-NAD cleavage-1 | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Babatunde, E, Dong, C.N, Xu, H.L, Henning, S. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
8OZ6
 
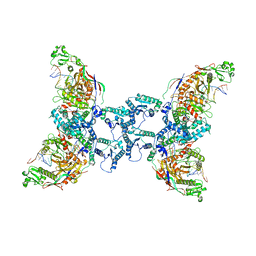 | | cryoEM structure of SPARTA complex ligand-free | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Piwi domain-containing protein, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Babatunde, E, Dong, C.N, Xu, H.L, Henning, S. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
8OZF
 
 | | cryoEM structure of SPARTA complex Tetramer Post-NAD cleavage-2 | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Piwi domain-containing protein, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Babatunde, E, Dong, C.N, Xu, H.L, Henning, S. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
2NLX
 
 | |
6QNK
 
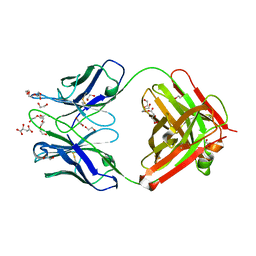 | | Antibody FAB fragment targeting Gi protein heterotrimer | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, FAB heavy chain, ... | | Authors: | Tsai, C.-J, Muehle, J, Pamula, F, Dawson, R.J.P, Maeda, S, Deupi, X, Schertler, G.F.X. | | Deposit date: | 2019-02-11 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryo-EM structure of the rhodopsin-G alpha i-beta gamma complex reveals binding of the rhodopsin C-terminal tail to the G beta subunit.
Elife, 8, 2019
|
|
8Q4H
 
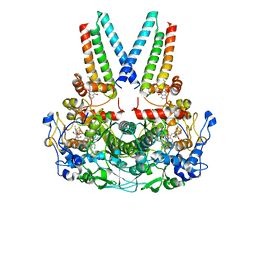 | | a membrane-bound menaquinol:organohalide oxidoreductase complex RDH complex | | Descriptor: | (~{Z})-1,2-bis(chloranyl)ethene, CO-METHYLCOBALAMIN, IRON/SULFUR CLUSTER, ... | | Authors: | Dongchun, N, Ekundayo, B, Henning, S, Julien, M, Holliger, C, Cimmino, L. | | Deposit date: | 2023-08-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structure of a membrane-bound menaquinol:organohalide oxidoreductase.
Nat Commun, 14, 2023
|
|
3FLR
 
 | |
3FLP
 
 | |
3FLT
 
 | |
7ZOA
 
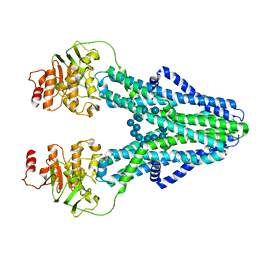 | | cryo-EM structure of CGT ABC transporter in presence of CBG substrate | | Descriptor: | Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, Cyclooctadecakis-(1-2)-(beta-D-glucopyranose) | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZO8
 
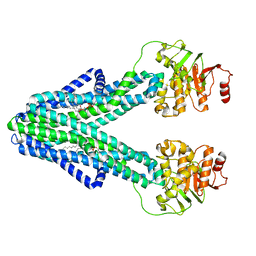 | | cryo-EM structure of CGT ABC transporter in nanodisc apo state | | Descriptor: | Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, DIUNDECYL PHOSPHATIDYL CHOLINE | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZO9
 
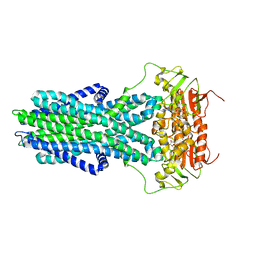 | | cryo-EM structure of CGT ABC transporter in vanadate trapped state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, VANADATE ION | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZNU
 
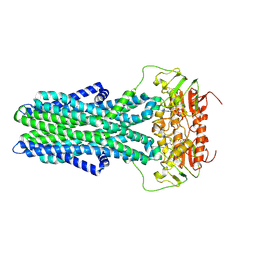 | | cryo-EM structure of CGT ABC transporter in detergent micelle | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, VANADATE ION | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-22 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZOL
 
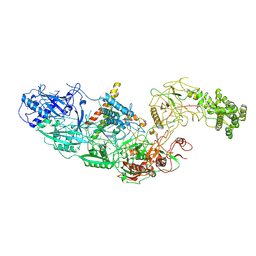 | | Cryo-EM structure of a CRISPR effector in complex with regulator | | Descriptor: | Cas7-11, TPR-CHAT, ZINC ION, ... | | Authors: | Babatunde, E.E, Davide, T, Bertrand, B, Sergey, N, Alexander, M, Henning, S, Dong, C.N. | | Deposit date: | 2022-04-26 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights into the regulation of Cas7-11 by TPR-CHAT.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZOQ
 
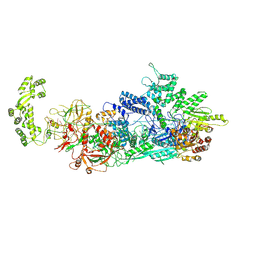 | | Cryo-EM structure of a CRISPR effector in complex with a caspase regulator | | Descriptor: | RNA (39-MER), TPR-CHAT, ZINC ION, ... | | Authors: | Babatunde, E.E, Davide, T, Bertrand, B, Sergey, N, Alexander, M, Henning, S, Dong, C.N. | | Deposit date: | 2022-04-26 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the regulation of Cas7-11 by TPR-CHAT.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6STS
 
 | | Human myelin protein P2 mutant R30Q | | Descriptor: | Myelin P2 protein, PALMITIC ACID, SULFATE ION | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2019-09-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6OGD
 
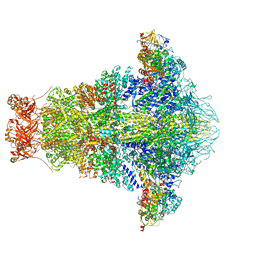 | | Cryo-EM structure of YenTcA in its prepore state | | Descriptor: | Chitinase 2, Toxin subunit YenA1, Toxin subunit YenA2 | | Authors: | Piper, S.J, Brillault, L, Box, J.K, Landsberg, M.J. | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures of the pore-forming A subunit from the Yersinia entomophaga ABC toxin.
Nat Commun, 10, 2019
|
|
6RIU
 
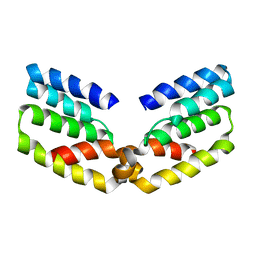 | |
6XUA
 
 | | Human myelin protein P2 mutant K21Q | | Descriptor: | CITRIC ACID, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XW9
 
 | | Human myelin protein P2 mutant K120S | | Descriptor: | CHLORIDE ION, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-23 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
5NIV
 
 | | Crystal structure of 5D3 Fab | | Descriptor: | Heavy chain of 5D3 Fab, Light chain of 5D3 Fab | | Authors: | Manolaridis, I, Locher, K.P. | | Deposit date: | 2017-03-27 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structure of the human multidrug transporter ABCG2.
Nature, 546, 2017
|
|
