2Y9W
 
 | | Crystal structure of PPO3, a tyrosinase from Agaricus bisporus, in deoxy-form that contains additional unknown lectin-like subunit | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, HOLMIUM ATOM, ... | | Authors: | Ismaya, W.T, Rozeboom, H.J, Weijn, A, Mes, J.J, Fusetti, F, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2011-02-17 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Agaricus Bisporus Mushroom Tyrosinase: Identity of the Tetramer Subunits and Interaction with Tropolone.
Biochemistry, 50, 2011
|
|
2Y9X
 
 | | Crystal structure of PPO3, a tyrosinase from Agaricus bisporus, in deoxy-form that contains additional unknown lectin-like subunit, with inhibitor tropolone | | Descriptor: | 2-HYDROXYCYCLOHEPTA-2,4,6-TRIEN-1-ONE, COPPER (II) ION, HOLMIUM ATOM, ... | | Authors: | Ismaya, W.T, Rozeboom, H.J, Weijn, A, Mes, J.J, Fusetti, F, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2011-02-17 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal Structure of Agaricus Bisporus Mushroom Tyrosinase: Identity of the Tetramer Subunits and Interaction with Tropolone.
Biochemistry, 50, 2011
|
|
3ZVH
 
 | | Methylaspartate ammonia lyase from Clostridium tetanomorphum mutant Q73A | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Raj, H, Szymanski, W, de Villiers, J, Rozeboom, H.J, Veetil, V.P, Reis, C.R, de Villiers, M, de Wildeman, S, Dekker, F.J, Quax, W.J, Thunnissen, A.M.W.H, Feringa, B.L, Janssen, D.B, Poelarends, G.J. | | Deposit date: | 2011-07-25 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering Methylaspartate Ammonia Lyase for the Asymmetric Synthesis of Unnatural Amino Acids.
Nat.Chem., 4, 2012
|
|
3ZVI
 
 | | Methylaspartate ammonia lyase from Clostridium tetanomorphum mutant L384A | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Raj, H, Szymanski, W, de Villiers, J, Rozeboom, H.J, Veetil, V.P, Reis, C.R, de Villiers, M, de Wildeman, S, Dekker, F.J, Quax, W.J, Thunnissen, A.M.W.H, Feringa, B.L, Janssen, D.B, Poelarends, G.J. | | Deposit date: | 2011-07-25 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering Methylaspartate Ammonia Lyase for the Asymmetric Synthesis of Unnatural Amino Acids.
Nat.Chem., 4, 2012
|
|
1A47
 
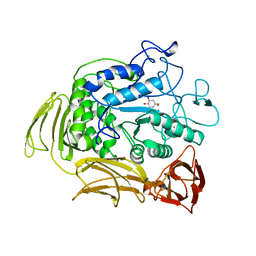 | | CGTASE FROM THERMOANAEROBACTERIUM THERMOSULFURIGENES EM1 IN COMPLEX WITH A MALTOHEXAOSE INHIBITOR | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Uitdehaag, J.C.M, Kalk, K.H, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1998-02-11 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Engineering of cyclodextrin product specificity and pH optima of the thermostable cyclodextrin glycosyltransferase from Thermoanaerobacterium thermosulfurigenes EM1.
J.Biol.Chem., 273, 1998
|
|
1QQ6
 
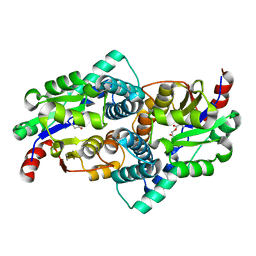 | | STRUCTURE OF L-2-HALOACID DEHALOGENASE FROM XANTHOBACTER AUTOTROPHICUS WITH CHLOROACETIC ACID COVALENTLY BOUND | | Descriptor: | CHLORIDE ION, PROTEIN (L-2-HALOACID DEHALOGENASE) | | Authors: | Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-06-11 | | Release date: | 1999-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of intermediates in the dehalogenation of haloalkanoates by L-2-haloacid dehalogenase.
J.Biol.Chem., 274, 1999
|
|
8QDH
 
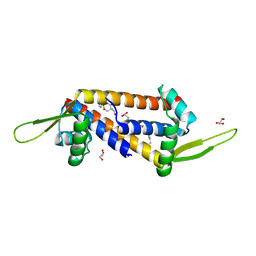 | | Engineered LmrR carrying a cyclic boronate ester formed between Tris and p-boronophenylalanine at position 89 | | Descriptor: | GLYCEROL, Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Rozeboom, H.J, Longwitz, L, Leveson-Gower, R.B, Roelfes, G. | | Deposit date: | 2023-08-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Boron catalysis in a designer enzyme.
Nature, 2024
|
|
8QDF
 
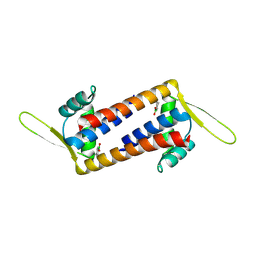 | | Engineered LmrR with Met-89 replaced by para-boronophenylalanine | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Rozeboom, H.J, Longwitz, L, Leveson-Gower, R.B, Roelfes, G. | | Deposit date: | 2023-08-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Boron catalysis in a designer enzyme.
Nature, 2024
|
|
1QQ7
 
 | | STRUCTURE OF L-2-HALOACID DEHALOGENASE FROM XANTHOBACTER AUTOTROPHICUS WITH CHLOROPROPIONIC ACID COVALENTLY BOUND | | Descriptor: | CHLORIDE ION, PROTEIN (L-2-HALOACID DEHALOGENASE) | | Authors: | Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-06-11 | | Release date: | 1999-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of intermediates in the dehalogenation of haloalkanoates by L-2-haloacid dehalogenase.
J.Biol.Chem., 274, 1999
|
|
1CDG
 
 | | NUCLEOTIDE SEQUENCE AND X-RAY STRUCTURE OF CYCLODEXTRIN GLYCOSYLTRANSFERASE FROM BACILLUS CIRCULANS STRAIN 251 IN A MALTOSE-DEPENDENT CRYSTAL FORM | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYL-TRANSFERASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Lawson, C.L, Van Montfort, R, Strokopytov, B.V, Kalk, K.H, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1993-08-02 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nucleotide sequence and X-ray structure of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 in a maltose-dependent crystal form.
J.Mol.Biol., 236, 1994
|
|
1QQ5
 
 | | STRUCTURE OF L-2-HALOACID DEHALOGENASE FROM XANTHOBACTER AUTOTROPHICUS | | Descriptor: | FORMIC ACID, PROTEIN (L-2-HALOACID DEHALOGENASE) | | Authors: | Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-06-10 | | Release date: | 1999-10-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of intermediates in the dehalogenation of haloalkanoates by L-2-haloacid dehalogenase.
J.Biol.Chem., 274, 1999
|
|
1SLY
 
 | | COMPLEX OF THE 70-KDA SOLUBLE LYTIC TRANSGLYCOSYLASE WITH BULGECIN A | | Descriptor: | 4-O-(4-O-SULFONYL-N-ACETYLGLUCOSAMININYL)-5-METHYLHYDROXY-L-PROLINE-TAURINE, 70-KDA SOLUBLE LYTIC TRANSGLYCOSYLASE | | Authors: | Thunnissen, A.M.W.H, Kalk, K.H, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1995-08-02 | | Release date: | 1996-08-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the 70-kDa soluble lytic transglycosylase complexed with bulgecin A. Implications for the enzymatic mechanism.
Biochemistry, 34, 1995
|
|
2XCZ
 
 | | Crystal Structure of macrophage migration inhibitory factor homologue from Prochlorococcus marinus | | Descriptor: | DI(HYDROXYETHYL)ETHER, POSSIBLE ATLS1-LIKE LIGHT-INDUCIBLE PROTEIN | | Authors: | Wasiel, A.A, Rozeboom, H.J, Hauke, D, Baas, B.J, Zandvoort, E, Quax, W.J, Thunnissen, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2010-04-27 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural and Functional Characterization of a Macrophage Migration Inhibitory Factor Homologue from the Marine Cyanobacterium Prochlorococcus Marinus.
Biochemistry, 49, 2010
|
|
1EHY
 
 | | X-ray structure of the epoxide hydrolase from agrobacterium radiobacter ad1 | | Descriptor: | POTASSIUM ION, PROTEIN (SOLUBLE EPOXIDE HYDROLASE) | | Authors: | Nardini, M, Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Rink, R, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 1998-10-17 | | Release date: | 1999-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The x-ray structure of epoxide hydrolase from Agrobacterium radiobacter AD1. An enzyme to detoxify harmful epoxides.
J.Biol.Chem., 274, 1999
|
|
1G4I
 
 | | Crystal structure of the bovine pancreatic phospholipase A2 at 0.97A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Steiner, R.A, Rozeboom, H.J, de Vries, A, Kalk, K.H, Murshudov, G.N, Wilson, K.S, Dijkstra, B.W. | | Deposit date: | 2000-10-27 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | X-ray structure of bovine pancreatic phospholipase A2 at atomic resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1LG1
 
 | |
1LG2
 
 | |
1LQ0
 
 | |
1PX0
 
 | | Crystal structure of the haloalcohol dehalogenase HheC complexed with the haloalcohol mimic (R)-1-para-nitro-phenyl-2-azido-ethanol | | Descriptor: | (R)-1-PARA-NITRO-PHENYL-2-AZIDO-ETHANOL, halohydrin dehalogenase | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Rozeboom, H.J, Kalk, K.H, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Mechanism of a Bacterial Haloalcohol Dehalogenase: a new variation of the short-chain dehydrogenase/reductase fold without an NAD(P)H binding site
EMBO J., 22, 2003
|
|
1PWX
 
 | | Crystal structure of the haloalcohol dehalogenase HheC complexed with bromide | | Descriptor: | BROMIDE ION, halohydrin dehalogenase | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Rozeboom, H.J, Kalk, K.H, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Mechanism of a Bacterial Haloalcohol Dehalogenase: a new variation of the short-chain dehydrogenase/reductase fold without an NAD(P)H binding site
EMBO J., 22, 2003
|
|
1PWZ
 
 | | Crystal structure of the haloalcohol dehalogenase HheC complexed with (R)-styrene oxide and chloride | | Descriptor: | CHLORIDE ION, R-STYRENE OXIDE, halohydrin dehalogenase | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Rozeboom, H.J, Kalk, K.H, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanism of a Bacterial Haloalcohol Dehalogenase: a new variation of the short-chain dehydrogenase/reductase fold without an NAD(P)H binding site
EMBO J., 22, 2003
|
|
1QBI
 
 | | SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE FROM ACINETOBACTER CALCOACETICUS | | Descriptor: | CALCIUM ION, GLYCEROL, PLATINUM (II) ION, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Kalk, K.H, Duine, J.A, Dijkstra, B.W. | | Deposit date: | 1999-04-22 | | Release date: | 2000-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The 1.7 A crystal structure of the apo form of the soluble quinoprotein glucose dehydrogenase from Acinetobacter calcoaceticus reveals a novel internal conserved sequence repeat.
J.Mol.Biol., 289, 1999
|
|
7ANV
 
 | |
2DIJ
 
 | | COMPLEX OF A Y195F MUTANT CGTASE FROM B. CIRCULANS STRAIN 251 COMPLEXED WITH A MALTONONAOSE INHIBITOR AT PH 9.8 OBTAINED AFTER SOAKING THE CRYSTAL WITH ACARBOSE AND MALTOHEXAOSE | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Strokopytov, B.V, Knegtel, R.M.A, Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 1998-05-27 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of cyclodextrin glycosyltransferase complexed with a maltononaose inhibitor at 2.6 angstrom resolution. Implications for product specificity.
Biochemistry, 35, 1996
|
|
2EDC
 
 | |
