6V79
 
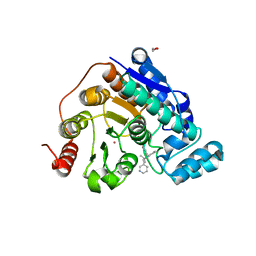 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with NF2376 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2S)-3,3-dimethyl-2-(pyridin-3-yl)-2,3-dihydro-1H-indol-1-yl]methyl}-N-hydroxybenzamide, Hdac6 protein, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03951526 Å) | | Cite: | Harnessing the Role of HDAC6 in Idiopathic Pulmonary Fibrosis: Design, Synthesis, Structural Analysis, and Biological Evaluation of Potent Inhibitors.
J.Med.Chem., 64, 2021
|
|
5LLF
 
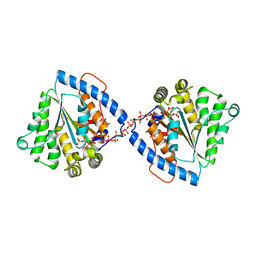 | |
5LDB
 
 | | Crystal Structure of Polyphosphate Kinase from Meiothermus ruber bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gerhardt, S, Einsle, O, Kemper, F, Schwarzer, N. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate recognition and mechanism revealed by ligand-bound polyphosphate kinase 2 structures.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
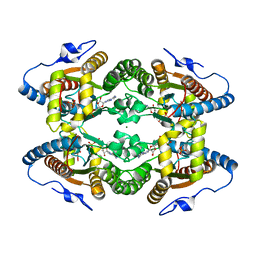
5LL0
 
 | |
5MAQ
 
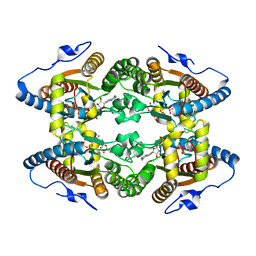 | | Crystal Structure of Polyphosphate Kinase from Meiothermus ruber bound to ADP and PPi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PYROPHOSPHATE, ... | | Authors: | Gerhardt, S, Einsle, O, Kemper, F, Schwarzer, N. | | Deposit date: | 2016-11-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Substrate recognition and mechanism revealed by ligand-bound polyphosphate kinase 2 structures.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
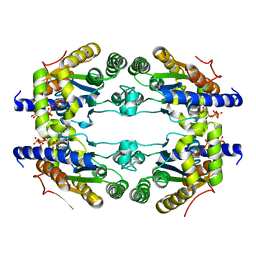
5NEU
 
 | |
6ST8
 
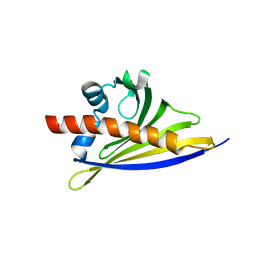 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein | | Descriptor: | Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
5LCD
 
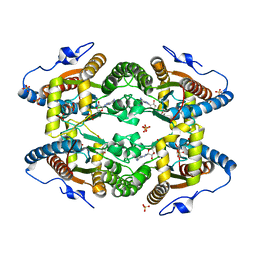 | | Structure of Polyphosphate Kinase from Meiothermus ruber bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gerhardt, S, Einsle, O, Kemper, F, Schwarzer, N. | | Deposit date: | 2016-06-21 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Substrate recognition and mechanism revealed by ligand-bound polyphosphate kinase 2 structures.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5LD1
 
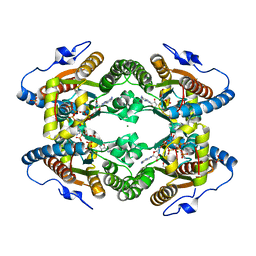 | | Crystal Structure of Polyphosphate Kinase from Meiothermus ruber bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Einsle, O, Kemper, F, Schwarzer, N. | | Deposit date: | 2016-06-23 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Substrate recognition and mechanism revealed by ligand-bound polyphosphate kinase 2 structures.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5NE4
 
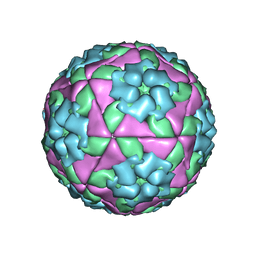 | |
5LLB
 
 | |
5NED
 
 | |
6STB
 
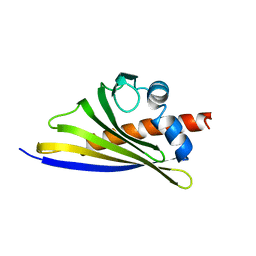 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein, Q64W mutant | | Descriptor: | Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
5LC9
 
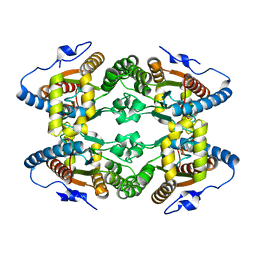 | |
6STA
 
 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein, E46A D48A mutant | | Descriptor: | Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
6ST9
 
 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein, D48R mutant | | Descriptor: | CHLORIDE ION, Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
5NER
 
 | |
7JSH
 
 | | Adeno-Associated Virus 2 Rep68 HD Heptamer-ssAAVS1 with ATPgS | | Descriptor: | DNA (5'-D(P*CP*GP*CP*TP*CP*GP*CP*TP*CP*GP*CP*TP*CP*GP*C)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
7JSG
 
 | | Adeno-Associated Virus 2 Rep68 HD-Heptamer-ssDNA with ATPgS | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
7JSI
 
 | | Adeno-Associated Virus 2 Rep68 HD Hexamer-ssDNA with ATPgS | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*T)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.01 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
7JSF
 
 | | Adeno-Associated Virus Helicase domain Heptamer with ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*T)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
7JSE
 
 | | Adeno-Associated Virus Origin Binding Domain in complex with ssDNA | | Descriptor: | DNA (5'-D(*TP*TP*TP*T)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
3PGS
 
 | | Phe3Gly mutant of EcFadL | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, ACETATE ION, CALCIUM ION, ... | | Authors: | van den Berg, B, Lepore, B.W. | | Deposit date: | 2010-11-02 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From the Cover: Ligand-gated diffusion across the bacterial outer membrane.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PIK
 
 | | Outer membrane protein CusC | | Descriptor: | Cation efflux system protein cusC, SULFATE ION, UNKNOWN LIGAND | | Authors: | van den Berg, B. | | Deposit date: | 2010-11-06 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli CusC, the Outer Membrane Component of a Heavy Metal Efflux Pump.
Plos One, 6, 2011
|
|
6V3P
 
 | |
