8F6T
 
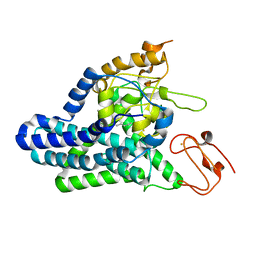 | | Cryo-EM structure of alkane 1-monooxygenase AlkB-AlkG complex from Fontimonas thermophila | | Descriptor: | Alkane 1-monooxygenase, DODECANE, FE (III) ION | | Authors: | Chai, J, Guo, G, McSweeney, S, Shanklin, J, Liu, Q. | | Deposit date: | 2022-11-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis for enzymatic terminal C-H bond functionalization of alkanes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2C6R
 
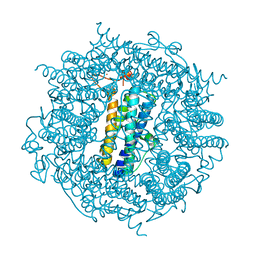 | | FE-SOAKED CRYSTAL STRUCTURE OF THE DPS92 FROM DEINOCOCCUS RADIODURANS | | Descriptor: | CHLORIDE ION, DNA-BINDING STRESS RESPONSE PROTEIN, DPS FAMILY, ... | | Authors: | Cuypers, M.G, Romao, C.V, Mitchell, E, Mcsweeney, S. | | Deposit date: | 2005-11-11 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Dps2 from Deinococcus Radiodurans Reveals an Unusual Pore Profile with a Non-Specific Metal Binding Site.
J.Mol.Biol., 371, 2007
|
|
4YTA
 
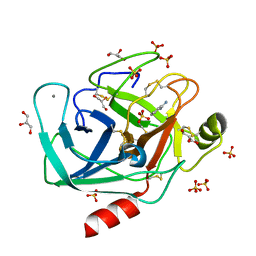 | | BOND LENGTH ANALYSIS OF ASP, GLU AND HIS RESIDUES IN TRYPSIN AT 1.2A RESOLUTION | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Fisher, S.J, Helliwell, J.R, Blakeley, M.P, Cianci, M, McSweeny, S. | | Deposit date: | 2015-03-17 | | Release date: | 2015-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation-state determination in proteins using high-resolution X-ray crystallography: effects of resolution and completeness.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
1DJL
 
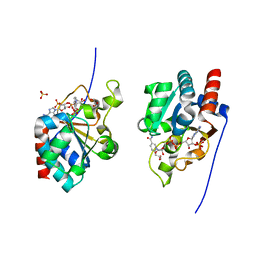 | | THE CRYSTAL STRUCTURE OF HUMAN TRANSHYDROGENASE DOMAIN III WITH BOUND NADP | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | White, S.A, Peak, S.J, Cotton, N.P, Jackson, J.B. | | Deposit date: | 1999-12-03 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The high-resolution structure of the NADP(H)-binding component (dIII) of proton-translocating transhydrogenase from human heart mitochondria.
Structure Fold.Des., 8, 2000
|
|
8G83
 
 | | Structure of NAD+ consuming protein Acinetobacter baumannii TIR domain | | Descriptor: | NAD(+) hydrolase AbTIR | | Authors: | Klontz, E.H, Wang, Y, Glendening, G, Carr, J, Tsibouris, T, Buddula, S, Nallar, S, Soares, A, Snyder, G.A. | | Deposit date: | 2023-02-17 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The structure of NAD + consuming protein Acinetobacter baumannii TIR domain shows unique kinetics and conformations.
J.Biol.Chem., 299, 2023
|
|
3DE1
 
 | |
3DDZ
 
 | |
3D9Q
 
 | |
3DE2
 
 | |
3DE0
 
 | |
3DE6
 
 | |
3DE3
 
 | |
3DE5
 
 | |
3DE7
 
 | |
3DE4
 
 | |
3DVS
 
 | |
4G2M
 
 | |
4G22
 
 | |
4G0B
 
 | |
5FXM
 
 | |
5WJH
 
 | | Using sound pulses to solve the crystal harvesting bottleneck | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CALCIUM ION, Proteinase K | | Authors: | Soares, A.S, Brennan, H.M, McCarthy, L, Leroy, L. | | Deposit date: | 2017-07-22 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Using sound pulses to solve the crystal-harvesting bottleneck.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
5WHW
 
 | | Using sound pulses to solve the crystal harvesting bottleneck | | Descriptor: | 1,2-ETHANEDIOL, BICINE, CALCIUM ION, ... | | Authors: | Soares, A.S, Brennan, H.M, Natarajan, R, McCarthy, L, Leroy, L. | | Deposit date: | 2017-07-18 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Using sound pulses to solve the crystal-harvesting bottleneck.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5WJG
 
 | | Using sound pulses to solve the crystal harvesting bottleneck | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, L(+)-TARTARIC ACID, ... | | Authors: | Soares, A.S, Brennan, H.M, McCarthy, L, Leroy, L. | | Deposit date: | 2017-07-22 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Using sound pulses to solve the crystal-harvesting bottleneck.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5FXL
 
 | |
