5AIH
 
 | | Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. CH55-sample-Native | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, LIMONENE-1,2-EPOXIDE HYDROLASE | | Authors: | Ferrandi, E, Sayer, C, Isupov, M.N, Annovazzi, C, Marchesi, C, Iacobone, G, Peng, X, Bonch-Osmolovskaya, E, Wohlgemuth, R, Littlechild, J.A, Montia, D. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries
FEBS J., 282, 2015
|
|
5AIF
 
 | | Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. Tomsk-sample-Native | | Descriptor: | IMIDAZOLE, LIMONENE-1,2-EPOXIDE HYDROLASE | | Authors: | Ferrandi, E, Sayer, C, Isupov, M.N, Annovazzi, C, Marchesi, C, Iacobone, G, Peng, X, Bonch-Osmolovskaya, E, Wohlgemuth, R, Littlechild, J.A, Montia, D. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries
FEBS J., 282, 2015
|
|
5AO9
 
 | | The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-native | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Sayer, C, Szabo, Z, Isupov, M.N, Ingham, C, Littlechild, J.A. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta Terrifontis Reveals an Open Active Site due to a Minimal 'CAP' Domain.
Front.Microbiol., 6, 2015
|
|
5AOC
 
 | | The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-valerate bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sayer, C, Szabo, Z, Isupov, M.N, Ingham, C, Littlechild, J.A. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta Terrifontis Reveals an Open Active Site due to a Minimal 'CAP' Domain.
Front.Microbiol., 6, 2015
|
|
6J4R
 
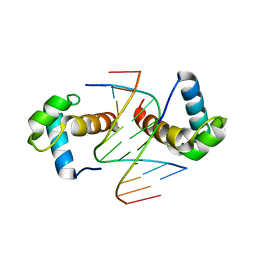 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*C)-3'), DNA (5'-D(*CP*CP*AP*TP*AP*TP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
6J5B
 
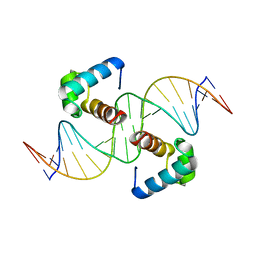 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*GP*GP*TP*AP*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*CP*AP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*TP*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*GP*TP*AP*CP*C)-3'), Protein PHOSPHATE STARVATION RESPONSE 1 | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N, Wu, Y.K. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
6I7S
 
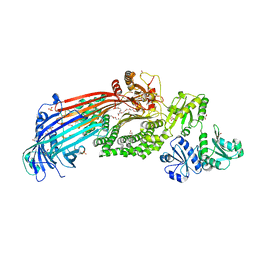 | | Microsomal triglyceride transfer protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Biterova, E, Isupov, M.N, Keegan, R.M, Lebedev, A.A, Ruddock, L.W. | | Deposit date: | 2018-11-17 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of human microsomal triglyceride transfer protein.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KXT
 
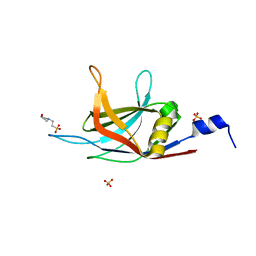 | | BON1-C2B | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Protein BONZAI 1, SULFATE ION | | Authors: | Wang, Q.C, Jiang, M.Q, Isupov, M.N, Sun, L.F, Wu, Y.K. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of an Arabidopsis Copine providing insights into this protein family
To be published
|
|
6KXU
 
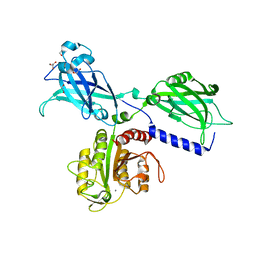 | | BON1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Q.C, Jiang, M.Q, Isupov, M.N, Sun, L.F, Wu, Y.K. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal Structure of an Arabidopsis Copine providing insights into this protein family
To be published
|
|
6KXK
 
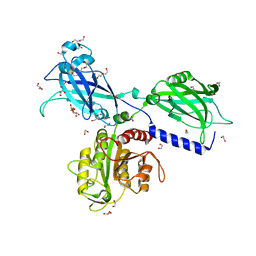 | | BON1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, Q.C, Jiang, M.Q, Isupov, M.N, Sun, L.F, Wu, Y.K. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Arabidopsis Copine providing insights into this protein family
To be published
|
|
8QQN
 
 | |
8QPQ
 
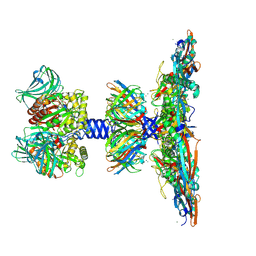 | | C1 turret to capsid interface of full Haloferax tailed virus 1 adjacent to the portal-capsid interface. | | Descriptor: | HK97 gp5-like major capsid protein, MAGNESIUM ION, Prokaryotic polysaccharide deacetylase, ... | | Authors: | Zhang, D, Daum, B, Isupov, M.N, McLaren, M. | | Deposit date: | 2023-10-02 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | CryoEM structure of Haloferax tailed virus 1
To Be Published
|
|
8QPG
 
 | | Turret of Haloferax tailed virus 1 | | Descriptor: | MAGNESIUM ION, Prokaryotic polysaccharide deacetylase, ZINC ION, ... | | Authors: | Zhang, D, Daum, B, Isupov, M.N, McLaren, M. | | Deposit date: | 2023-10-02 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | CryoEM structure of Haloferax tailed virus 1
To Be Published
|
|
2YN4
 
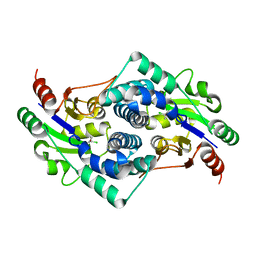 | | L-2-chlorobutryic acid bound complex L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | (2S)-2-chlorobutanoic acid, L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-12 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2YMM
 
 | | Sulfate bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE, SULFATE ION | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-09 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
7NVJ
 
 | | Crystal structure of UFC1 Y110A & F121A | | Descriptor: | GLYCEROL, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
7NVK
 
 | | Crystal structure of UBA5 fragment fused to the N-terminus of UFC1 | | Descriptor: | Ubiquitin-like modifier-activating enzyme 5,Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
7NW1
 
 | | Crystal structure of UFC1 in complex with UBA5 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
2WS2
 
 | |
7NZQ
 
 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-mannose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7NZO
 
 | | D-lyxose isomerasefrom the hyperthermophilic archaeon Thermofilum sp | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7NZP
 
 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-fructose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
2YMP
 
 | | Chloroacetic acid complex bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2YMQ
 
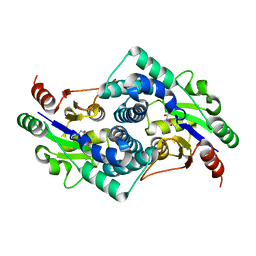 | | Chloropropionic acid complex bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2YML
 
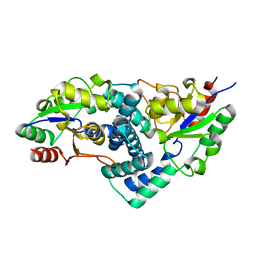 | | Native L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-09 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
