1O5H
 
 | |
1O3U
 
 | |
1O22
 
 | |
1O4V
 
 | |
2KYS
 
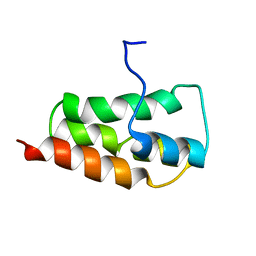 | | NMR Structure of the SARS Coronavirus Nonstructural Protein Nsp7 in Solution at pH 6.5 | | Descriptor: | Non-structural protein 7 | | Authors: | Johnson, M.A, Jaudzems, K, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the SARS-CoV Nonstructural Protein 7 in Solution at pH 6.5.
J.Mol.Biol., 402, 2010
|
|
1O1Z
 
 | |
1O2D
 
 | |
1O4U
 
 | |
1O4W
 
 | |
1O20
 
 | |
1O4S
 
 | |
2JZD
 
 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2HSX
 
 | | NMR Structure of the nonstructural protein 1 (nsp1) from the SARS coronavirus | | Descriptor: | Leader protein; p65 homolog; NSP1 (EC 3.4.22.-) | | Authors: | Almeida, M.S, Herrmann, T, Geralt, M, Johnson, M.A, Saikatendu, K, Joseph, J, Subramanian, R.C, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-07-24 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2JZE
 
 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3, single conformer closest to the mean coordinates of an ensemble of twenty energy minimized conformers | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
1O58
 
 | |
1O5U
 
 | |
2JZF
 
 | | NMR Conformer closest to the mean coordinates of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
1TYP
 
 | |
1EO8
 
 | | INFLUENZA VIRUS HEMAGGLUTININ COMPLEXED WITH A NEUTRALIZING ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY (HEAVY CHAIN), ... | | Authors: | Fleury, D, Gigant, B, Daniels, R.S, Skehel, J.J, Knossow, M, Bizebard, T. | | Deposit date: | 2000-03-22 | | Release date: | 2000-04-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural evidence for recognition of a single epitope by two distinct antibodies.
Proteins, 40, 2000
|
|
1KB5
 
 | | MURINE T-CELL RECEPTOR VARIABLE DOMAIN/FAB COMPLEX | | Descriptor: | ANTIBODY DESIRE-1, KB5-C20 T-CELL ANTIGEN RECEPTOR | | Authors: | Housset, D, Mazza, G, Gregoire, C, Piras, C, Malissen, B, Fontecilla-Camps, J.C. | | Deposit date: | 1997-04-06 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The three-dimensional structure of a T-cell antigen receptor V alpha V beta heterodimer reveals a novel arrangement of the V beta domain.
EMBO J., 16, 1997
|
|
1KVF
 
 | |
1KVG
 
 | | EPO-3 beta Hairpin Peptide | | Descriptor: | Protein: EPO-3 Receptor Agonist | | Authors: | Skelton, N.J, Russell, S, de Sauvage, F, Cochran, A.G. | | Deposit date: | 2002-01-25 | | Release date: | 2002-03-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Amino Acid Determinants of beta-Hairpin Conformation in Erythropoeitin
Receptor Agonist Peptides Derived from a Phage Display Library
J.Mol.Biol., 316, 2002
|
|
1NAM
 
 | | MURINE ALLOREACTIVE SCFV TCR-PEPTIDE-MHC CLASS I MOLECULE COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BM3.3 T Cell Receptor alpha-Chain, BM3.3 T Cell Receptor beta-Chain, ... | | Authors: | Reiser, J.-B, Darnault, C, Gregoire, C, Mosser, T, Mazza, G, Kearnay, A, van der Merwe, P.A, Fontecilla-Camps, J.C, Housset, D, Malissen, B. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CDR3 loop flexibility contributes to the degeneracy of TCR recognition
Nat.Immunol., 4, 2003
|
|
1QB6
 
 | | BOVINE TRYPSIN 3,3'-[3,5-DIFLUORO-4-METHYL-2, 6-PYRIDINEDIYLBIS(OXY)]BIS(BENZENECARBOXIMIDAMIDE) (ZK-805623) COMPLEX | | Descriptor: | 3,3'-[3,5-DIFLUORO-4-METHYL-2,6-PYRIDYLENEBIS(OXY)]-BIS(BENZENECARBOXIMIDAMIDE), CALCIUM ION, POTASSIUM ION, ... | | Authors: | Whitlow, M. | | Deposit date: | 1999-04-29 | | Release date: | 2000-04-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic analysis of potent and selective factor Xa inhibitors complexed to bovine trypsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1QBO
 
 | |
