1V5B
 
 | | The Structure Of The Mutant, S225A and E251L, Of 3-Isopropylmalate Dehydrogenase From Bacillus Coagulans | | Descriptor: | 3-isopropylmalate dehydrogenase, SULFATE ION | | Authors: | Fujita, K, Minami, H, Suzuki, K, Tsunoda, M, Sekiguchi, T, Mizui, R, Tsuzaki, S, Nakamura, S, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of a highly thermo-stabilized mutant of 3-isopropylmalate dehydrogenase from Bacillus coagulans: An evaluation of local packing density in the hydrophobic core
To be Published
|
|
1V3N
 
 | | Crystal structure of d(GCGAGAGC): the DNA quadruplex structure split from the octaplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*AP*GP*AP*GP*C)-3', POTASSIUM ION | | Authors: | Kondo, J, Umeda, S, Sunami, T, Takenaka, A. | | Deposit date: | 2003-11-03 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats
Nucleic Acids Res., 32, 2004
|
|
1V5F
 
 | | Crystal Structure of Pyruvate oxidase complexed with FAD and TPP, from Aerococcus viridans | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, Pyruvate oxidase, ... | | Authors: | Hossain, M.T, Suzuki, K, Yamamoto, T, Imamura, S, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of pyruvate oxidase from Aerococcus viridans with cofactors and with a reaction intermediate reveal the flexibility of the active-site tunnel for catalysis.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1VAY
 
 | | Crystal Structure of Uricase from Arthrobacter globiformis with inhibitor 8-azaxanthine | | Descriptor: | 8-AZAXANTHINE, Uric acid oxidase | | Authors: | Hossain, M.T, Suzuki, K, Yamamoto, T, Imamura, S, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2004-02-19 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of Uricase from Arthrobacter Globiformis Complexed with an inhibitor 8-Azaxanthine
To be Published
|
|
1V53
 
 | | The crystal structure of 3-isopropylmalate dehydrogenase from Bacillus coagulans | | Descriptor: | 3-isopropylmalate dehydrogenase | | Authors: | Fujita, K, Minami, H, Suzuki, K, Tsunoda, M, Sekiguchi, T, Mizui, R, Tsuzaki, S, Nakamura, S, Takenaka, A. | | Deposit date: | 2003-11-20 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The crystal structure of 3-isopropylmalate dehydrogenase from Bacillus coagulans
To be Published
|
|
1V5E
 
 | | Crystal Structure of Pyruvate oxidase containing FAD, from Aerococcus viridans | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyruvate oxidase, SULFATE ION | | Authors: | Hossain, M.T, Suzuki, K, Yamamoto, T, Imamura, S, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Pyruvate oxidase containing FAD, from Aerococcus viridans
To be Published
|
|
1UHY
 
 | | Crystal structure of d(GCGATAGC): the base-intercalated duplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*AP*TP*AP*GP*C)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Kondo, J, Umeda, S.I, Fujita, K, Sunami, T, Takenaka, A. | | Deposit date: | 2003-07-13 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray analyses of d(GCGAXAGC) containing G and T at X: the base-intercalated duplex is still stable even in point mutants at the fifth residue.
J.Synchrotron Radiat., 11, 2004
|
|
2YQU
 
 | | Crystal structures and evolutionary relationship of two different lipoamide dehydrogenase(E3s) from Thermus thermophilus | | Descriptor: | 2-oxoglutarate dehydrogenase E3 component, CARBONATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kondo, H, Hossain, M.T, Adachi, W, Nakai, T, Kamiya, N, Kuramitsu, K. | | Deposit date: | 2007-03-31 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures and evolutionary relationship of two different lipoamide dehydrogenase(E3s) from Thermus thermophilus
To be Published
|
|
1XER
 
 | | STRUCTURE OF FERREDOXIN | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, ZINC ION | | Authors: | Fujii, T, Hata, Y, Moriyama, H, Wakagi, T, Tanaka, N, Oshima, T. | | Deposit date: | 1996-08-28 | | Release date: | 1997-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding centre in thermoacidophilic archaeal ferredoxins.
Nat.Struct.Biol., 3, 1996
|
|
1IPD
 
 | | THREE-DIMENSIONAL STRUCTURE OF A HIGHLY THERMOSTABLE ENZYME, 3-ISOPROPYLMALATE DEHYDROGENASE OF THERMUS THERMOPHILUS AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, SULFATE ION | | Authors: | Imada, K, Sato, M, Tanaka, N, Katsube, Y, Matsuura, Y, Oshima, T. | | Deposit date: | 1992-01-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 A resolution.
J.Mol.Biol., 222, 1991
|
|
2CMM
 
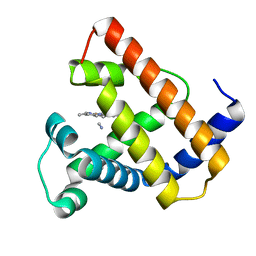 | | STRUCTURAL ANALYSIS OF THE MYOGLOBIN RECONSTITUTED WITH IRON PORPHINE | | Descriptor: | CYANIDE ION, MYOGLOBIN, PORPHYRIN FE(III) | | Authors: | Sato, T, Tanaka, N, Moriyama, H, Igarashi, N, Neya, S, Funasaki, N, Iizuka, T, Shiro, Y. | | Deposit date: | 1993-12-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the myoglobin reconstituted with iron porphine.
J.Biol.Chem., 268, 1993
|
|
