8VK9
 
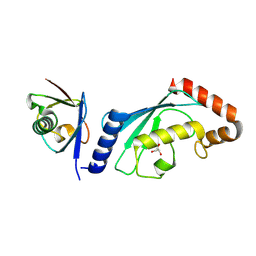 | | Structure of UbV.d2.3 in complex with Ube2d2-S22R | | Descriptor: | GLYCEROL, Ubiquitin variant D2.3, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Middleton, A.J. | | Deposit date: | 2024-01-08 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biophysical characterisation of ubiquitin variants that inhibit the ubiquitin conjugating enzyme Ube2d2
To Be Published
|
|
8SJQ
 
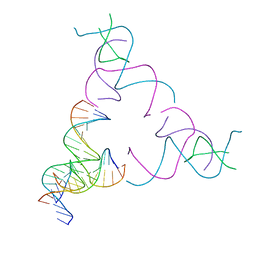 | | [3T16] Self-assembling right-handed tensegrity triangle with 16 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*GP*CP*CP*TP*GP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*CP*TP*GP*TP*GP*GP*CP*AP*TP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (6.19 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJM
 
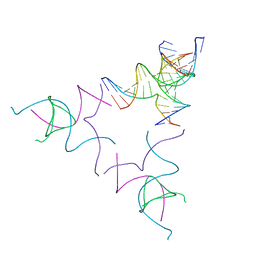 | | [3T12] Self-assembling left-handed tensegrity triangle with 12 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*CP*GP*CP*CP*TP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*GP*CP*AP*TP*GP*TP*GP*GP*CP*GP*AP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*AP*TP*GP*CP*GP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (8.08 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJR
 
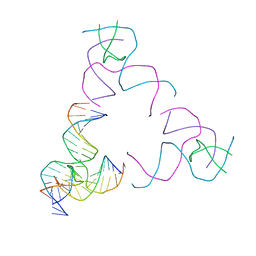 | | [3T17] Self-assembling right-handed tensegrity triangle with 17 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*GP*CP*CP*TP*GP*AP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*CP*TP*GP*TP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*TP*AP*TP*TP*CP*AP*CP*CP*AP*CP*GP*AP*T)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (5.25 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJU
 
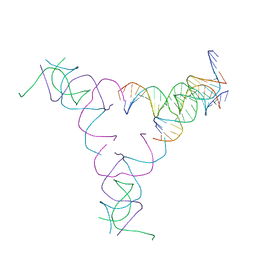 | | [4T17] Self-assembling right-handed four-turn tensegrity triangle with 17 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (25-MER), DNA (5'-D(*GP*AP*AP*AP*AP*AP*CP*AP*CP*TP*GP*CP*CP*TP*GP*AP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(P*GP*CP*GP*GP*TP*AP*TP*TP*CP*AP*CP*CP*AP*CP*GP*AP*T)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.14 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJO
 
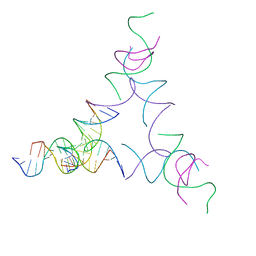 | | [3T14] Self-assembling left-handed tensegrity triangle with 14 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(P*CP*AP*CP*GP*TP*GP*GP*AP*CP*AP*GP*GP*AP*G)-3'), DNA (5'-D(P*CP*AP*GP*CP*TP*CP*AP*GP*CP*CP*TP*GP*AP*CP*TP*CP*A)-3'), DNA (5'-D(P*GP*TP*GP*AP*GP*TP*CP*TP*CP*CP*AP*CP*GP*T)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.08 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJN
 
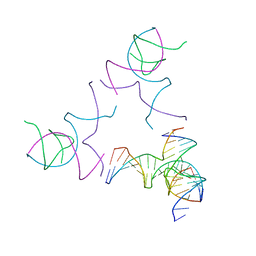 | | [3T13] Self-assembling left-handed tensegrity triangle with 13 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*CP*GP*CP*CP*TP*GP*AP*CP*TP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*GP*CP*TP*GP*TP*GP*GP*CP*GP*AP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*GP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.3 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJP
 
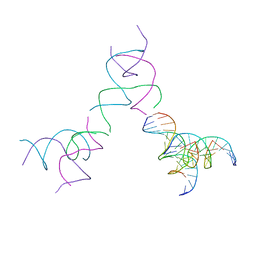 | | [3T15] Self-assembling DNA motif with 15 base pairs between junctions and P32 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*TP*GP*AP*CP*CP*TP*GP*AP*CP*TP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*CP*TP*GP*TP*GP*GP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*CP*GP*AP*TP*GP*GP*AP*CP*AP*GP*GP*GP*G)-3'), ... | | Authors: | Vecchioni, S, Janowski, J, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (5.22 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJS
 
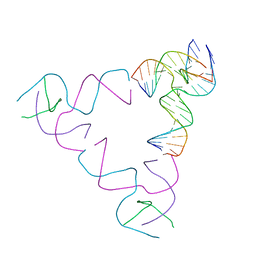 | | [3T18] Self-assembling right-handed tensegrity triangle with 18 interjunction base pairs and P63 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*GP*CP*CP*TP*GP*AP*CP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*CP*TP*GP*TP*GP*GP*CP*TP*C)-3'), DNA (5'-D(P*TP*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (6.31 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJW
 
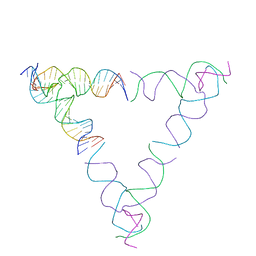 | | [4T28] Self-assembling right-handed tensegrity triangle with 28 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (28-MER), DNA (5'-D(*GP*AP*AP*CP*TP*GP*CP*CP*TP*GP*AP*AP*TP*TP*AP*CP*TP*GP*AP*CP*CP*G)-3'), DNA (5'-D(*TP*CP*AP*TP*CP*AP*GP*TP*GP*GP*CP*AP*GP*T)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.64 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SL5
 
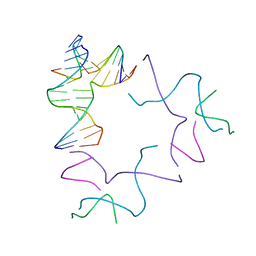 | | [2T13] Self-assembling left-handed two-turn tensegrity triangle with 13 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*CP*TP*GP*AP*CP*TP*AP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*TP*GP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*A)-3'), ... | | Authors: | Vecchioni, S, Janowski, J, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (6.55 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJT
 
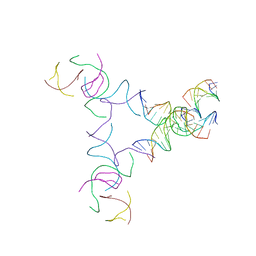 | | [3T14+10] Self-assembling left-handed tensegrity triangle with 14 interjunction base pairs and a 10 bp linker with R3 symmetry | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*CP*CP*TP*GP*AP*GP*GP*TP*CP*GP*AP*GP*C)-3'), DNA (5'-D(*GP*AP*CP*TP*CP*TP*GP*CP*TP*A)-3'), DNA (5'-D(*GP*TP*TP*AP*GP*CP*AP*GP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (9.38 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJV
 
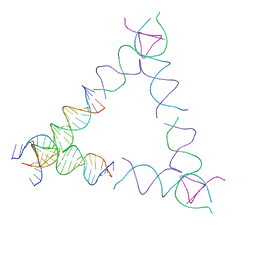 | | [4T24] Self-assembling left-handed tensegrity triangle with 24 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(P*CP*TP*TP*GP*TP*AP*GP*TP*CP*TP*CP*AP*CP*CP*AP*CP*TP*GP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*GP*AP*AP*CP*AP*CP*TP*CP*CP*TP*GP*AP*GP*AP*CP*TP*AP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*CP*AP*TP*CP*AP*CP*AP*GP*TP*GP*GP*AP*CP*TP*AP*CP*AP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (8.59 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SZ5
 
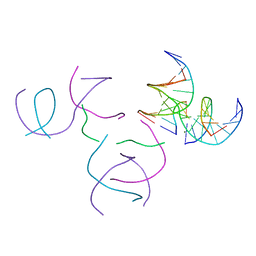 | | [2T5] Self-assembling DNA motif with 5 base pairs between junctions and P32 symmetry | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*G)-3'), DNA (5'-D(P*AP*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(P*CP*AP*CP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Janowski, J, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-05-26 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5ZTY
 
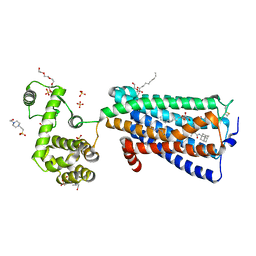 | | Crystal structure of human G protein coupled receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, X.T, Hua, T, Wu, L.J, Liu, Z.J. | | Deposit date: | 2018-05-05 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Human Cannabinoid Receptor CB2
Cell, 176, 2019
|
|
7SPL
 
 | | [2T3] Self-assembling 3D DNA triangle with three inter-junction base pairs containing the L1 junction and a zero-linked center strand | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, DNA (5'-D(*GP*AP*C)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*TP*CP*C)-3'), ... | | Authors: | Vecchioni, S, Lu, B, Sha, R, Ohayon, Y.P, Seeman, N.C. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.09 Å) | | Cite: | The Rule of Thirds: Controlling Junction Chirality and Polarity in 3D DNA Tiles.
Small, 19, 2023
|
|
7U3R
 
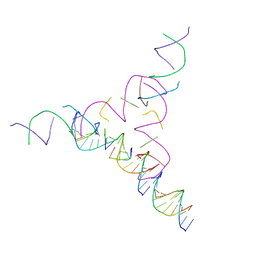 | | [2T7+10] Self-assembling tensegrity triangle with two turns of DNA and the sticky end attachment of a one-turn linker per axis, with R3 symmetry | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*GP*AP*TP*GP*CP*TP*GP*AP*GP*T)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Lu, B, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.27 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3Y
 
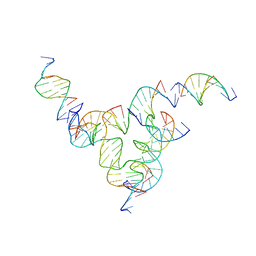 | | [L233] Self-assembling tensegrity triangle with two turns, three turns and three turns of DNA per axis by linker addition with P1 symmetry | | Descriptor: | DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*AP*CP*TP*GP*AP*TP*GP*TP*GP*GP*TP*AP*GP*G)-3'), DNA (5'-D(*AP*GP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.06 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
8D93
 
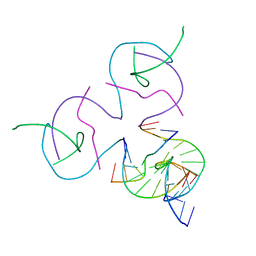 | | [2T7] Self-assembling tensegrity triangle with R3 symmetry at 2.96 A resolution, update and junction cut for entry 3GBI | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*TP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*AP*CP*CP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Woloszyn, K, Lu, B, Sha, R, Ohayon, Y.P, Seeman, N.C. | | Deposit date: | 2022-06-09 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The Rule of Thirds: Controlling Junction Chirality and Polarity in 3D DNA Tiles.
Small, 19, 2023
|
|
7U3Q
 
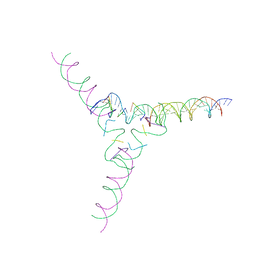 | | [4T7] Self-assembling tensegrity triangle with four turns of DNA per axis with R3 symmetry | | Descriptor: | DNA (29-MER), DNA (42-MER), DNA (5'-D(P*AP*CP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Lu, B, Ma, Y, Seeman, N.C, Sha, R, Ohayon, Y.P, Huang, Q. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (9.32 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3W
 
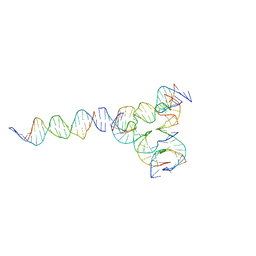 | | [L224] Self-assembling tensegrity triangle with two turns, two turns and four turns of DNA per axis by linker addition with P1 symmetry | | Descriptor: | DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*AP*GP*TP*CP*GP*TP*GP*GP*CP*TP*CP*G)-3'), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.33 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3Z
 
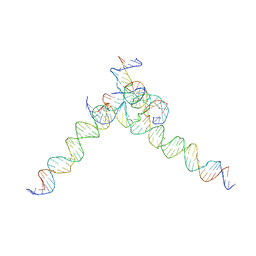 | | [F244] Self-assembling tensegrity triangle with two turns, four turns and four turns of DNA per axis by extension with P1 symmetry | | Descriptor: | DNA (35-MER), DNA (42-MER), DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.55 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3V
 
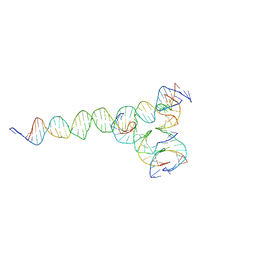 | | [F224] Self-assembling tensegrity triangle with two turns, two turns and four turns of DNA per axis by extension with P1 symmetry | | Descriptor: | DNA (35-MER), DNA (42-MER), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (5.14 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U41
 
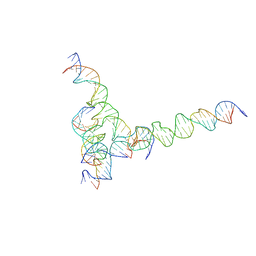 | | [F234] Self-assembling tensegrity triangle with two turns, three turns and four turns of DNA per axis by extension with P1 symmetry | | Descriptor: | DNA (31-MER), DNA (35-MER), DNA (42-MER), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.24 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U42
 
 | | [F334] Self-assembling tensegrity triangle with three turns, three turns and four turns of DNA per axis by extension with P1 symmetry | | Descriptor: | DNA (31-MER), DNA (35-MER), DNA (42-MER), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.71 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
