6T5I
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with inhibitor of WNT production (IWP)-2 | | Descriptor: | DIMETHYL SULFOXIDE, Listeriolysin positive regulatory factor A, SODIUM ION, ... | | Authors: | Oelker, M, Grundstrom, C, Blumenthal, A, Sauer-Eriksson, A.E. | | Deposit date: | 2019-10-16 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of the master regulator of Listeria monocytogenes virulence enables bacterial clearance from spacious replication vacuoles in infected macrophages.
Plos Pathog., 18, 2022
|
|
1LLS
 
 | | CRYSTAL STRUCTURE OF UNLIGANDED MALTOSE BINDING PROTEIN WITH XENON | | Descriptor: | Maltose-binding periplasmic protein, XENON | | Authors: | Rubin, S.M, Lee, S.-Y, Ruiz, E.J, Pines, A, Wemmer, D.E. | | Deposit date: | 2002-04-30 | | Release date: | 2002-09-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DETECTION AND CHARACTERIZATION OF XENON-BINDING SITES IN PROTEINS BY 129XE NMR SPECTROSCOPY
J.MOL.BIOL., 322, 2002
|
|
1MDP
 
 | |
1OMP
 
 | |
3NCA
 
 | | X-ray structure of ketohexokinase in complex with a thieno pyridinol compound | | Descriptor: | Ketohexokinase, SULFATE ION, thieno[3,2-b]pyridin-7-ol | | Authors: | Abad, M.C, Gibbs, A.C, Spurlino, J.C. | | Deposit date: | 2010-06-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Electron density guided fragment-based lead discovery of ketohexokinase inhibitors.
J.Med.Chem., 53, 2010
|
|
3NBW
 
 | | X-ray structure of ketohexokinase in complex with a pyrazole compound | | Descriptor: | 5-amino-3-(methylsulfanyl)-1-phenyl-1H-pyrazole-4-carbonitrile, GLYCEROL, Ketohexokinase, ... | | Authors: | Abad, M.C, Gibbs, A.C, Spurlino, J.C. | | Deposit date: | 2010-06-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Electron density guided fragment-based lead discovery of ketohexokinase inhibitors.
J.Med.Chem., 53, 2010
|
|
3NC2
 
 | |
3NC9
 
 | |
3SHI
 
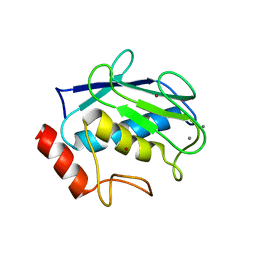 | | Crystal structure of human MMP1 catalytic domain at 2.2 A resolution | | Descriptor: | CALCIUM ION, Interstitial collagenase, ZINC ION | | Authors: | Bertini, I, Calderone, V, Cerofolini, L, Fragai, M, Geraldes, C.F.G.C, Hermann, P, Luchinat, C, Parigi, G, Teixeira, J. | | Deposit date: | 2011-06-16 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The catalytic domain of MMP-1 studied through tagged lanthanides.
Febs Lett., 586, 2012
|
|
2I0V
 
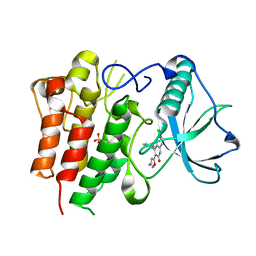 | | c-FMS tyrosine kinase in complex with a quinolone inhibitor | | Descriptor: | 6-CHLORO-3-(3-METHYLISOXAZOL-5-YL)-4-PHENYLQUINOLIN-2(1H)-ONE, SULFATE ION, cFMS tyrosine kinase | | Authors: | Schubert, C, Schalk-Hihi, C. | | Deposit date: | 2006-08-11 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the tyrosine kinase domain of colony-stimulating factor-1 receptor (cFMS) in complex with two inhibitors.
J.Biol.Chem., 282, 2007
|
|
2I0Y
 
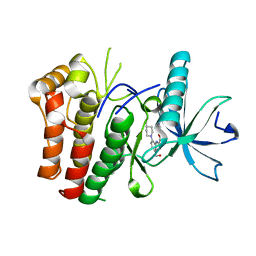 | | cFMS tyrosine kinase (FGF KID) in complex with an arylamide inhibitor | | Descriptor: | 5-CYANO-FURAN-2-CARBOXYLIC ACID [5-HYDROXYMETHYL-2-(4-METHYL-PIPERIDIN-1-YL)-PHENYL]-AMIDE, cFMS Tyrosine Kinase | | Authors: | Schubert, C, Schalk-Hihi, C. | | Deposit date: | 2006-08-11 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the tyrosine kinase domain of colony-stimulating factor-1 receptor (cFMS) in complex with two inhibitors.
J.Biol.Chem., 282, 2007
|
|
2I1M
 
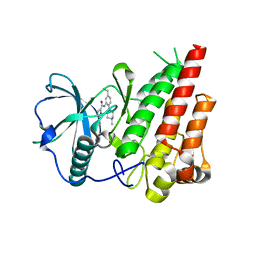 | | cFMS tyrosine kinase (tie2 KID) in complex with an arylamide inhibitor | | Descriptor: | 5-CYANO-FURAN-2-CARBOXYLIC ACID [5-HYDROXYMETHYL-2-(4-METHYL-PIPERIDIN-1-YL)-PHENYL]-AMIDE, Macrophage colony-stimulating factor 1 receptor | | Authors: | Schubert, C, Schalk-Hihi, C. | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the tyrosine kinase domain of colony-stimulating factor-1 receptor (cFMS) in complex with two inhibitors.
J.Biol.Chem., 282, 2007
|
|
