6AFO
 
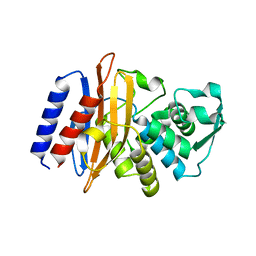 | |
5DBU
 
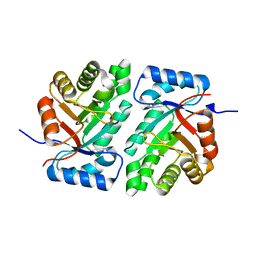 | |
5H6H
 
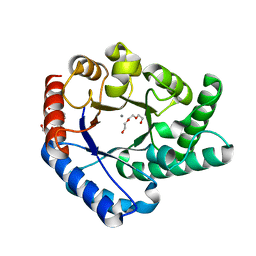 | | Crystal Structure of Hyperthermophilic Thermotoga maritima L-Ribulose 3-Epimerase with Mn2+ | | Descriptor: | MANGANESE (II) ION, PENTAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | Authors: | Cao, T.-P, Choi, J.M, Shin, S.M, Le, D.W, Lee, S.H. | | Deposit date: | 2016-11-13 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | TM0416, a Hyperthermophilic Promiscuous Nonphosphorylated Sugar Isomerase, Catalyzes Various C5and C6Epimerization Reactions
Appl. Environ. Microbiol., 83, 2017
|
|
5DBT
 
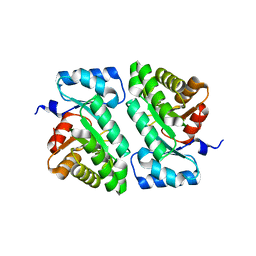 | |
7X89
 
 | | Tid1 | | Descriptor: | DnaJ homolog subfamily A member 3, mitochondrial | | Authors: | Jang, J, Lee, S.H, Kang, D.H, Sim, D.W, Jo, K.S, Ryu, H, Kim, E.H, Ryu, K.S, Lee, J.H, Kim, J.H, Won, H.S. | | Deposit date: | 2022-03-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the J-domain and GF-motif of the mitochondrial Hsp40, Tid1
To Be Published
|
|
6J4N
 
 | | Structure of papua new guinea MBL-1(PNGM-1) native | | Descriptor: | Metallo-beta-lactamases PNGM-1, ZINC ION | | Authors: | Hong, M.K, Park, K.S, Jeon, J.H, Lee, J.H, Park, Y.S, Lee, S.H, Kang, L.W. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The novel metallo-beta-lactamase PNGM-1 from a deep-sea sediment metagenome: crystallization and X-ray crystallographic analysis.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3U0R
 
 | | Helical repeat structure of apoptosis inhibitor 5 reveals protein-protein interaction modules | | Descriptor: | Apoptosis inhibitor 5 | | Authors: | Han, B.G, Kim, K.H, Jeong, K.C, Cho, J.W, Noh, K.H, Kim, T.W, Yoon, H.J, Suh, S.W, Lee, S.H, Lee, B.I. | | Deposit date: | 2011-09-29 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Helical repeat structure of apoptosis inhibitor 5 reveals protein-protein interaction modules.
J.Biol.Chem., 287, 2012
|
|
7CDH
 
 | | Crystal structure of Betaaspartyl dipeptidase from thermophilic keratin degrading Fervidobacterium islandicum-AW-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Isoaspartyl dipeptidase, ... | | Authors: | Dhanasingh, I, La, J.W, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional Characterization of Primordial Protein Repair Enzyme M38 Metallo-Peptidase From Fervidobacterium islandicum AW-1.
Front Mol Biosci, 7, 2020
|
|
7CWV
 
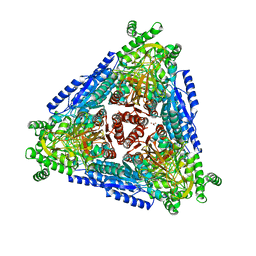 | | Crystal structure of Arabinose isomerase from hyper thermophilic bacterium Thermotoga maritima (TMAI) wt | | Descriptor: | GLYCEROL, L-arabinose isomerase, MANGANESE (II) ION | | Authors: | Hoang, N.K.Q, Dhanasingh, I, Cao, T.P, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-08-31 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Crystal structure of Arabinose isomerase from hyper thermophilic bacterium Thermotoga maritima (TMAI) wt
To Be Published
|
|
7CX7
 
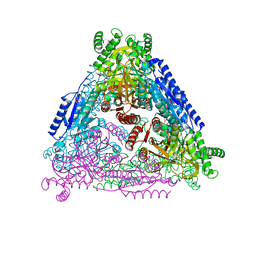 | | Crystal structure of Arabinose isomerase from hybrid AI8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, L-arabinose isomerase, ... | | Authors: | Hoang, N.K.Q, Dhanasingh, I, Cao, T.P, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of Arabinose isomerase from hyper thermophilic bacterium Thermotoga maritima (TMAI) wt
To Be Published
|
|
7CH3
 
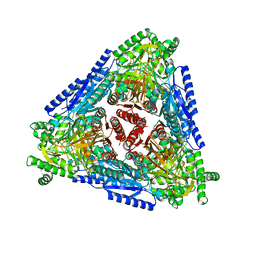 | | Crystal structure of Arabinose isomerase from hyper thermophilic bacterium Thermotoga maritima (TMAI) triple mutant (K264A, E265A, K266A) | | Descriptor: | L-arabinose isomerase, MANGANESE (II) ION | | Authors: | Cao, T.P, Dhanasingh, I, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-07-04 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Crystal structure of Arabinose isomerase from hyper thermophilic bacterium Thermotoga maritima (TMAI) triple mutant (K264A, E265A, K266A)
To Be Published
|
|
7CHL
 
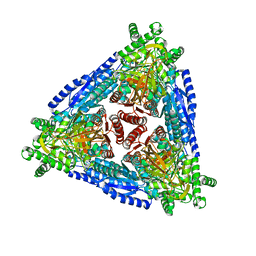 | | Crystal structure of hybrid Arabinose isomerase AI-10 | | Descriptor: | Hybrid Arabinose isomerase, MANGANESE (II) ION, SODIUM ION | | Authors: | Cao, T.P, Dhanasingh, I, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-07-06 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of hybrid Arabinose isomerase AI-10
To Be Published
|
|
7CXO
 
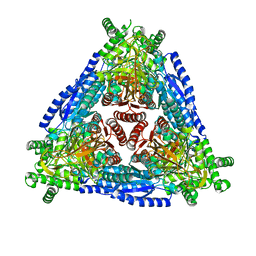 | | Crystal structure of Arabinose isomerase from hybrid AI10 | | Descriptor: | GLYCEROL, L-arabinose isomerase, MANGANESE (II) ION | | Authors: | Hoang, N.K.Q, Dhanasingh, I, Cao, T.P, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-09-02 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Arabinose isomerase from hyper thermophilic hybrid AI10
To Be Published
|
|
7CYY
 
 | | Crystal structure of Arabinose isomerase from hybrid AI8 with Adonitol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, D-ribitol, L-arabinose isomerase, ... | | Authors: | Hoang, N.K.Q, Dhanasingh, I, Cao, T.P, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-09-05 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Arabinose isomerase from hyper thermophilic hybrid AI8 with Adonitol
To Be Published
|
|
7CF6
 
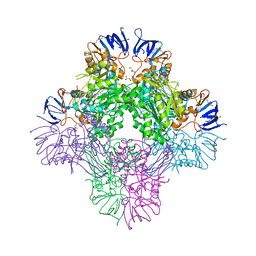 | | Crystal structure of Beta-aspartyl dipeptidase from thermophilic keratin degrading Fervidobacterium islandicum AW-1 in complex with beta-Asp-Leu dipeptide | | Descriptor: | (2S)-2-[[(3S)-3-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]amino]-4-methyl-pentanoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Dhanasingh, I, La, J.W, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Functional Characterization of Primordial Protein Repair Enzyme M38 Metallo-Peptidase From Fervidobacterium islandicum AW-1.
Front Mol Biosci, 7, 2020
|
|
1ZM1
 
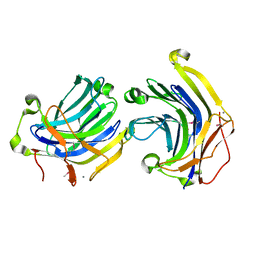 | | Crystal structures of complex F. succinogenes 1,3-1,4-beta-D-glucanase and beta-1,3-1,4-cellotriose | | Descriptor: | Beta-glucanase, CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Tsai, L.C, Shyur, L.F, Cheng, Y.S, Lee, S.H. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase in complex with beta-1,3-1,4-cellotriose
J.Mol.Biol., 354, 2005
|
|
6JKW
 
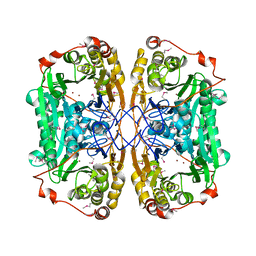 | | Seleno-methionine PNGM-1 from deep-sea sediment metagenome | | Descriptor: | Metallo-beta-lactamases PNGM-1, ZINC ION | | Authors: | Hong, M.K, Park, K.S, Jeon, J.H, Lee, J.H, Park, Y.S, Lee, S.H, Kang, L.W. | | Deposit date: | 2019-03-02 | | Release date: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | PNGM 1 a novel subclass B3 metallo beta lactamase from a deep sea sediment metagenome
Journal of Global Antimicrobial Resistance, 14, 2018
|
|
7D28
 
 | |
7D2Q
 
 | |
7D2P
 
 | |
7D2M
 
 | |
7D2N
 
 | |
5XM3
 
 | | Crystal Structure of Methanol dehydrogenase from Methylophaga aminisulfidivorans | | Descriptor: | Glucose dehydrogenase, MAGNESIUM ION, Methanol dehydrogenase [cytochrome c] subunit 2, ... | | Authors: | Cao, T.P, Choi, J.M, Lee, S.H. | | Deposit date: | 2017-05-12 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The crystal structure of methanol dehydrogenase, a quinoprotein from the marine methylotrophic bacterium Methylophaga aminisulfidivorans MPT
J. Microbiol., 56, 2018
|
|
5WUJ
 
 | | Crystal structure of FliF-FliG complex from H. pylori | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, GLYCEROL | | Authors: | Au, S.W, Xue, C, Lam, K.H, Lee, S.H. | | Deposit date: | 2016-12-19 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the FliF-FliG complex from Helicobacter pylori yields insight into the assembly of the motor MS-C ring in the bacterial flagellum
J. Biol. Chem., 293, 2018
|
|
4NTZ
 
 | | Crystal structure of Adenylate kinase from Streptococcus pneumoniae | | Descriptor: | Adenylate kinase, SULFATE ION | | Authors: | Thach, T.T, Luong, T.T, Lee, S.H, Rhee, D.K. | | Deposit date: | 2013-12-03 | | Release date: | 2014-07-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Adenylate kinase from Streptococcus pneumoniae is essential for growth through its catalytic activity
FEBS Open Bio, 4, 2014
|
|
