5FA0
 
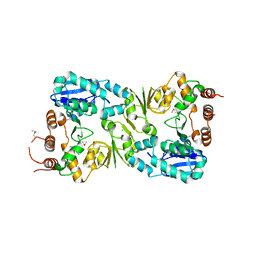 | | The structure of the beta-3-deoxy-D-manno-oct-2-ulosonic acid transferase domain from WbbB | | Descriptor: | CHLORIDE ION, Putative N-acetyl glucosaminyl transferase | | Authors: | Mallette, E, Ovchinnikova, O.G, Whitfield, C, Kimber, M.S. | | Deposit date: | 2015-12-10 | | Release date: | 2016-05-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bacterial beta-Kdo glycosyltransferases represent a new glycosyltransferase family (GT99).
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
3GGP
 
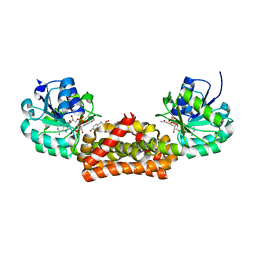 | | Crystal structure of prephenate dehydrogenase from A. aeolicus in complex with hydroxyphenyl propionate and NAD+ | | Descriptor: | CHLORIDE ION, HYDROXYPHENYL PROPIONIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sun, W, Shahinas, D, Kimber, M.S, Christendat, D. | | Deposit date: | 2009-03-01 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Crystal Structure of Aquifex aeolicus Prephenate Dehydrogenase Reveals the Mode of Tyrosine Inhibition.
J.Biol.Chem., 284, 2009
|
|
3SSQ
 
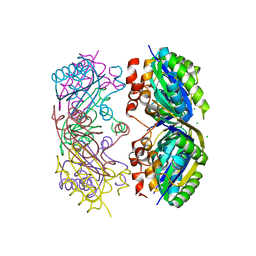 | | CcmK2 - form 1 dodecamer | | Descriptor: | CHLORIDE ION, Carbon dioxide concentrating mechanism protein, GLYCEROL | | Authors: | Samborska, B, Kimber, M.S. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A CcmK2 double layer is the dominant architectural feature of the beta-carboxysomal shell facet
Structure, 2012
|
|
6MGD
 
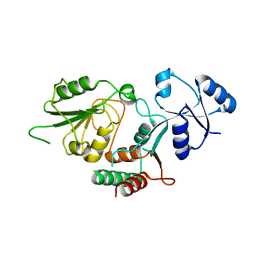 | | Thermosulfurimonas dismutans KpsC, beta Kdo 2,7 transferase | | Descriptor: | Capsular polysaccharide export system protein KpsC | | Authors: | Doyle, L, Mallette, E, Kimber, M.S, Whitfield, C. | | Deposit date: | 2018-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biosynthesis of a conserved glycolipid anchor for Gram-negative bacterial capsules.
Nat.Chem.Biol., 15, 2019
|
|
6MGB
 
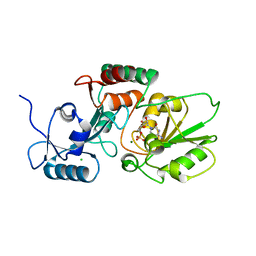 | | Thermosulfurimonas dismutans KpsC, beta Kdo 2,4 transferase | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, Capsular polysaccharide export system protein KpsC, ... | | Authors: | Doyle, L, Mallette, E, Kimber, M.S, Whitfield, C. | | Deposit date: | 2018-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biosynthesis of a conserved glycolipid anchor for Gram-negative bacterial capsules.
Nat.Chem.Biol., 15, 2019
|
|
6MR1
 
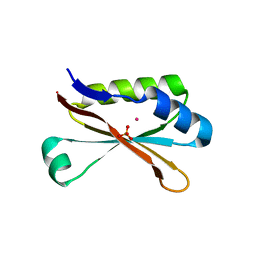 | | RbcS-like subdomain of CcmM | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Carbon dioxide concentrating mechanism protein, ... | | Authors: | Ryan, P, Kimber, M.S. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The small RbcS-like domains of the beta-carboxysome structural protein CcmM bind RubisCO at a site distinct from that binding the RbcS subunit.
J. Biol. Chem., 294, 2019
|
|
6MGC
 
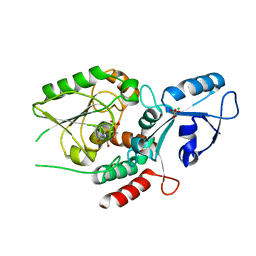 | | Escherichia coli KpsC, N-terminal domain | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, Capsule polysaccharide export protein KpsC, ... | | Authors: | Doyle, L, Mallette, E, Kimber, M.S. | | Deposit date: | 2018-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Biosynthesis of a conserved glycolipid anchor for Gram-negative bacterial capsules.
Nat.Chem.Biol., 15, 2019
|
|
5SUH
 
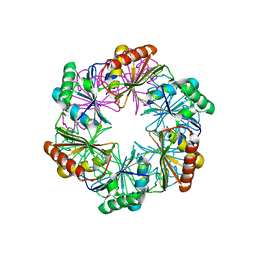 | |
2NWQ
 
 | | Short chain dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | Probable short-chain dehydrogenase, SULFATE ION | | Authors: | McGrath, T.E, Kimber, M.S, Beattie, B, Thambipillai, D, Dharamsi, A, Virag, C, Nethery-Brokx, K, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2006-11-16 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Short chain dehydrogenase from Pseudomonas aeruginosa
To be Published
|
|
3Q9O
 
 | |
2Q6M
 
 | |
2Q5T
 
 | | Full-length Cholix toxin from Vibrio Cholerae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cholix toxin | | Authors: | Jorgensen, R, Fieldhouse, R.J, Merrill, A.R. | | Deposit date: | 2007-06-01 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cholix Toxin, a Novel ADP-ribosylating Factor from Vibrio cholerae.
J.Biol.Chem., 283, 2008
|
|
3GGO
 
 | | Crystal structure of prephenate dehydrogenase from A. aeolicus with HPP and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Prephenate dehydrogenase | | Authors: | Sun, W, Shahinas, D, Christendat, D. | | Deposit date: | 2009-03-01 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Aquifex aeolicus Prephenate Dehydrogenase Reveals the Mode of Tyrosine Inhibition.
J.Biol.Chem., 284, 2009
|
|
3GGG
 
 | |
4FXQ
 
 | | Full-length Certhrax toxin from Bacillus cereus in complex with Inhibitor P6 | | Descriptor: | 8-fluoro-2-(3-piperidin-1-ylpropanoyl)-1,3,4,5-tetrahydrobenzo[c][1,6]naphthyridin-6(2H)-one, CHLORIDE ION, Putative ADP-ribosyltransferase Certhrax, ... | | Authors: | Visschedyk, D.D, Dimov, S, Kimber, M.S, Park, H.W, Merrill, A.R. | | Deposit date: | 2012-07-03 | | Release date: | 2012-09-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9599 Å) | | Cite: | Certhrax Toxin, an Anthrax-related ADP-ribosyltransferase from Bacillus cereus.
J.Biol.Chem., 287, 2012
|
|
4FML
 
 | |
