7NYM
 
 | | Mutant V517A - SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | HEXAETHYLENE GLYCOL, PHOSPHATE ION, SH3 domain of JNK-interacting Protein 1 (JIP1), ... | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.614 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NZB
 
 | | Mutant V517L of the SH3 domain of JNK-interacting protein 1 (JIP1) | | Descriptor: | PHOSPHATE ION, SH3 domain of JNK-interacting protein 1 (JIP1), TETRAETHYLENE GLYCOL | | Authors: | Perez, L.M, Ielasi, F.S, Jensen, M.R, Palencia, A. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NYN
 
 | | Mutant Y526A of SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.537 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
1MZI
 
 | | Solution ensemble structures of HIV-1 gp41 2F5 mAb epitope | | Descriptor: | 2F5 epitope of HIV-1 gp41 fusion protein | | Authors: | Barbato, G, Bianchi, E, Ingallinella, P, Hurni, W.H, Miller, M.D, Ciliberto, G, Cortese, R, Bazzo, R, Shiver, J.W, Pessi, A. | | Deposit date: | 2002-10-08 | | Release date: | 2003-09-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the epitope of the anti-HIV antibody 2F5 sheds light into its mechanism of neutralization and HIV fusion.
J.Mol.Biol., 330, 2003
|
|
8RMP
 
 | | Influenza polymerase A/H7N9-4M (ENDO(R) | Core1) | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RN3
 
 | |
8RN5
 
 | |
8RMQ
 
 | | Influenza polymerase A/H7N9-4M (ENDO(R) | Core2) | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RMR
 
 | |
8RN7
 
 | |
8RN8
 
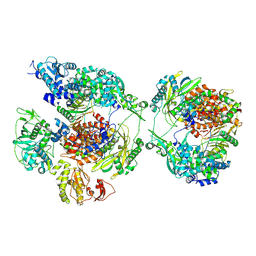 | | Influenza B polymerase pseudo-symmetrical apo-dimer (FluPol(E)|FluPol(S)) | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RNB
 
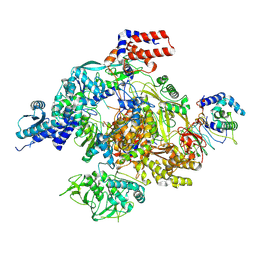 | |
8RNC
 
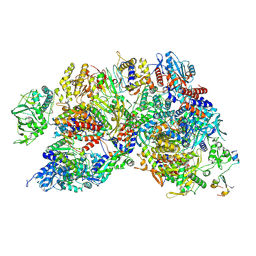 | | Influenza B polymerase, replication complex, an asymmetric polymerase dimer bound to human ANP32A (from "Influenza B polymerase apo-trimer" | Local refinement) | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member A, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RMS
 
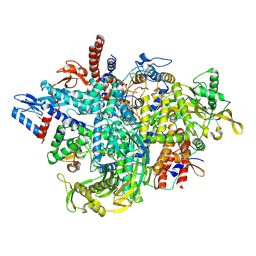 | |
8RN0
 
 | |
8RN1
 
 | | Influenza B polymerase, monomeric encapsidase with 5' cRNA hook bound | | Descriptor: | 5' cRNA hook (1-12), Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RN2
 
 | | Monomeric apo-influenza B polymerase, encapsidase conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RN4
 
 | |
8RN6
 
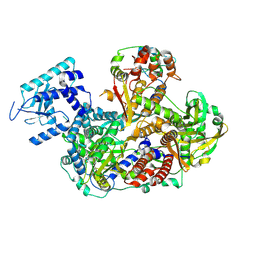 | |
8RN9
 
 | |
8RNA
 
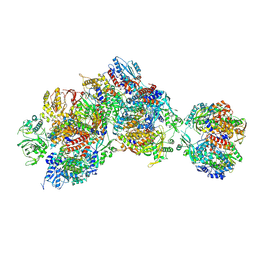 | | Influenza B polymerase apo-trimer | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member A, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
2KRN
 
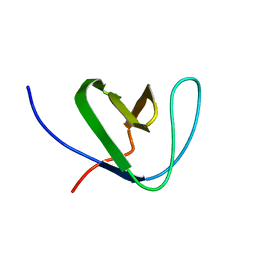 | |
