5ZZZ
 
 | | The crystal structure of Mandelate oxidase Y128C with benzoyl-formic acid | | Descriptor: | 4-hydroxymandelate oxidase, BENZOYL-FORMIC ACID, FLAVIN MONONUCLEOTIDE | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | The flavin mononucleotide cofactor in alpha-hydroxyacid oxidases exerts its electrophilic/nucleophilic duality in control of the substrate-oxidation level.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6A21
 
 | | Mandelate oxidase mutant-Y128F with the N5-malonyl-FMN adduct | | Descriptor: | 3-[7,8-dimethyl-2,4-bis(oxidanylidene)-10-[(2S,3S,4R)-2,3,4-tris(oxidanyl)-5-phosphonooxy-pentyl]-1H-benzo[g]pteridin-5-yl]-3-oxidanylidene-propanoic acid, 4-hydroxymandelate oxidase | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | The flavin mononucleotide cofactor in alpha-hydroxyacid oxidases exerts its electrophilic/nucleophilic duality in control of the substrate-oxidation level.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5ZZQ
 
 | | The crystal structure of Mandelate oxidase with (S)-4-Hydroxymandelic acid | | Descriptor: | (2S)-hydroxy(4-hydroxyphenyl)ethanoic acid, 4-hydroxymandelate oxidase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-04 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Biochemical and structural explorations of alpha-hydroxyacid oxidases reveal a four-electron oxidative decarboxylation reaction.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6A4H
 
 | | Mandelate oxidase mutant-Y128F with the peroxide FMN adduct | | Descriptor: | 4-hydroxymandelate oxidase, [(2~{R},3~{S},4~{S})-5-[(4~{a}~{S})-4~{a}-(dioxidanyl)-7,8-dimethyl-2,4-bis(oxidanylidene)-5~{H}-benzo[g]pteridin-10-yl]-2,3,4-tris(oxidanyl)pentyl] dihydrogen phosphate | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-19 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and chemical trapping of flavin-oxide intermediates reveals substrate-directed reaction multiplicity.
Protein Sci., 29, 2020
|
|
6A11
 
 | | Mandelate oxidase mutant-Y128F with phenylpyruvic acid | | Descriptor: | 3-PHENYLPYRUVIC ACID, 4-hydroxymandelate oxidase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical and structural explorations of alpha-hydroxyacid oxidases reveal a four-electron oxidative decarboxylation reaction.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6A1B
 
 | | Mandelate oxidase mutant-Y128F with 3,3,3-trifluoro-2,2-dihydroxypropanoic acid | | Descriptor: | 3,3,3-trifluoro-2,2-dihydroxypropanoic acid, 4-hydroxymandelate oxidase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural and chemical trapping of flavin-oxide intermediates reveals substrate-directed reaction multiplicity.
Protein Sci., 29, 2020
|
|
6A1M
 
 | | Mandelate oxidase mutant-Y128F with 5-deazariboflavin mononucleotide and benzoylformic acid | | Descriptor: | 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-5-O-phosphono-D-ribitol, 4-hydroxymandelate oxidase, BENZOYL-FORMIC ACID, ... | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The flavin mononucleotide cofactor in alpha-hydroxyacid oxidases exerts its electrophilic/nucleophilic duality in control of the substrate-oxidation level.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6A4G
 
 | | Mandelate oxidase mutant-Y128F with the monooxide FMN adduct | | Descriptor: | 1-deoxy-1-[(4aS)-4a-hydroxy-7,8-dimethyl-2,4-dioxo-3,4,4a,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono-D-ribitol, 4-hydroxymandelate oxidase | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-19 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and chemical trapping of flavin-oxide intermediates reveals substrate-directed reaction multiplicity.
Protein Sci., 29, 2020
|
|
5ZZX
 
 | |
6A00
 
 | | The crystal structure of Mandelate oxidase with (S)-2-phenylpropionate | | Descriptor: | (2~{S})-2-phenylpropanoic acid, 4-hydroxymandelate oxidase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biochemical and structural explorations of alpha-hydroxyacid oxidases reveal a four-electron oxidative decarboxylation reaction.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4OCJ
 
 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, SODIUM ION | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCU
 
 | | N-acetylhexosamine 1-phosphate kinase_ATCC15697 in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, SULFATE ION | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCO
 
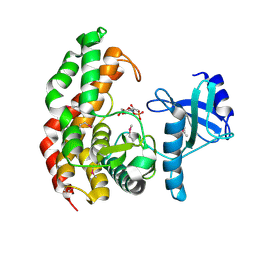 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc-1-phosphate | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCQ
 
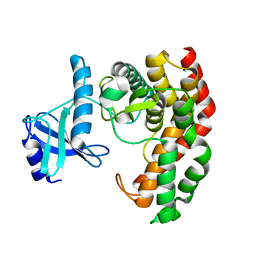 | | N-acetylhexosamine 1-phosphate kinase in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, N-acetylhexosamine 1-phosphate kinase | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCP
 
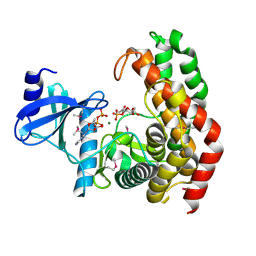 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc-1-phosphate and ADP | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCK
 
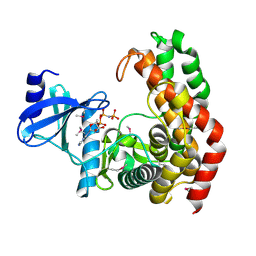 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc and AMPPNP | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCV
 
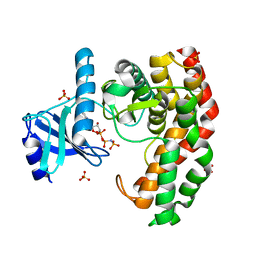 | | N-acetylhexosamine 1-phosphate kinase_ATCC15697 in complex with GlcNAc and AMPPNP | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6IIX
 
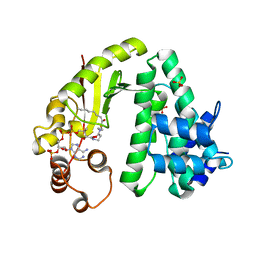 | |
6M2U
 
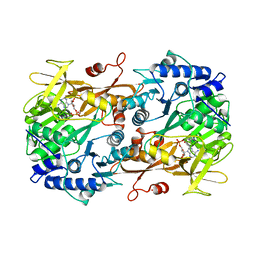 | |
6M2O
 
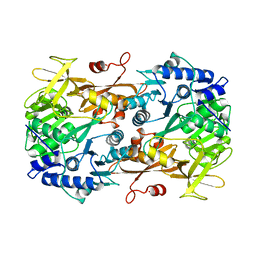 | |
5HGX
 
 | |
5HJE
 
 | |
5HYV
 
 | |
5I4I
 
 | |
5I5G
 
 | |
