6UHH
 
 | | Crystal Structure of Human RYR Receptor 3 ( 848-1055) in Complex with ATP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, R, Kim, Y, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-09-27 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.138 Å) | | Cite: | Crystal Structure of Human RYR Receptor 3 ( 848-1055) in Complex with ATP
To Be Published
|
|
6UHA
 
 | |
2LNC
 
 | | Solution NMR structure of Norwalk virus protease | | Descriptor: | 3C-like protease | | Authors: | Takahashi, D, Hiromasa, Y, Kim, Y, Anbanandam, A, Chang, K, Prakash, O. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamics characterization of norovirus protease.
Protein Sci., 22, 2013
|
|
7L91
 
 | | Structure of Metallo Beta-Lactamase L1 in a Complex with Hydrolyzed Moxalactam Determined by Pink-Beam Serial Crystallography | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Vukica, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-01 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
6V72
 
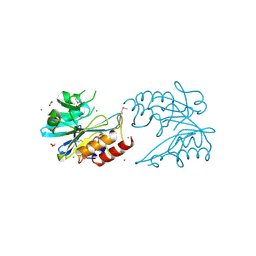 | | Crystal Structure of Metallo Beta Lactamase from Erythrobacter litoralis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase II, CALCIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Erythrobacter litoralis
To Be Published
|
|
4FK1
 
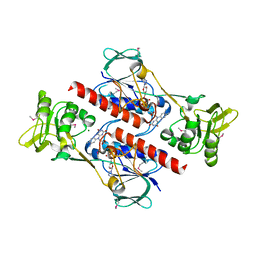 | | Crystal Structure of Putative Thioredoxin Reductase TrxB from Bacillus anthracis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-12 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Crystal Structure of Putative Thioredoxin Reductase TrxB from Bacillus anthracis
To be Published
|
|
6V70
 
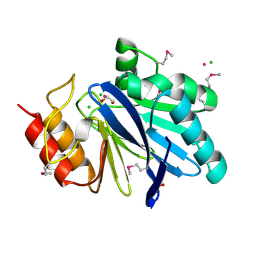 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Cadmium in the Active Site | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CADMIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica.
To Be Published
|
|
7M02
 
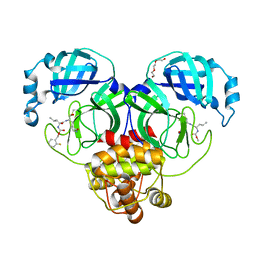 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 17c | | Descriptor: | (1R,2S)-2-((S)-2-((((2-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((2-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZW
 
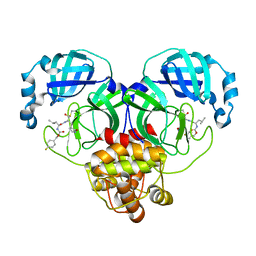 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 20b (deuterated analog of 19b) | | Descriptor: | (1R,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZY
 
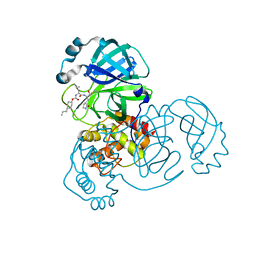 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 3c | | Descriptor: | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZZ
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 5c | | Descriptor: | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1r,4S)-4-phenylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1r,4S)-4-phenylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7M03
 
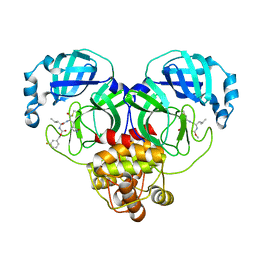 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 18c | | Descriptor: | (1R,2S)-2-((S)-2-((((3-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((3-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7M01
 
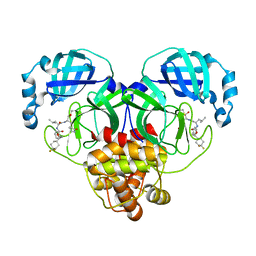 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 14c | | Descriptor: | (1R,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)-2-phenylethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)-2-phenylethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZT
 
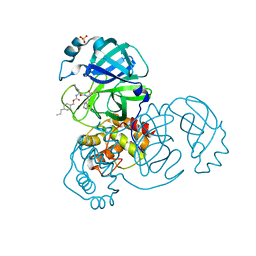 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 8b | | Descriptor: | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)methoxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)methoxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZX
 
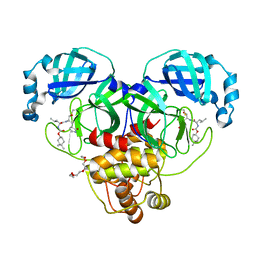 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 1c | | Descriptor: | (1R,2S)-2-((S)-2-((((4,4-dimethylcyclohexyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((4,4-dimethylcyclohexyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZU
 
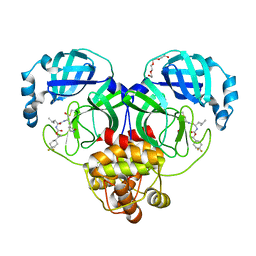 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 12b | | Descriptor: | (1R,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)ethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)ethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7M04
 
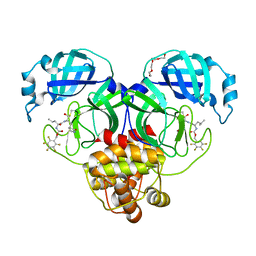 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 21c | | Descriptor: | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-((((perfluorophenyl)methoxy)carbonyl)amino)pentanamido)-3-((R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl)propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-((((perfluorophenyl)methoxy)carbonyl)amino)pentanamido)-3-((R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZV
 
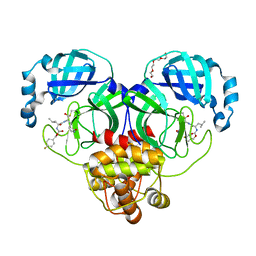 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 19b | | Descriptor: | (1R,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7M00
 
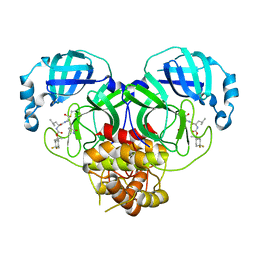 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 13c | | Descriptor: | (1R,2S)-2-((S)-2-((((2-(4,4-difluorocyclohexyl)propan-2-yl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((2-(4,4-difluorocyclohexyl)propan-2-yl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
6V54
 
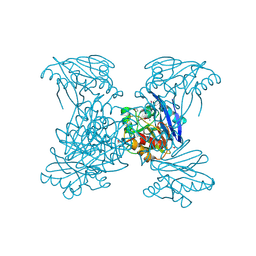 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-03 | | Release date: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica.
To Be Published
|
|
6VC6
 
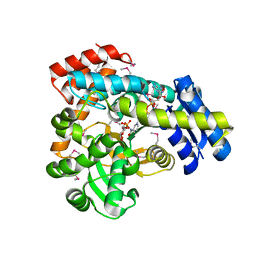 | | 2.1 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Mn2+ | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-phospho-alpha-glucosidase, GLYCEROL, ... | | Authors: | Wu, R, Kim, Y, Endres, M, Joachimiak, J. | | Deposit date: | 2019-12-20 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Mn2+
To Be Published
|
|
6VH3
 
 | | 2.20 A resolution structure of MERS 3CL protease in complex with inhibitor 7j | | Descriptor: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6V61
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in the Complex with the Inhibitor Captopril | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in the Complex with the Inhibitor Captopril.
To Be Published
|
|
6VH2
 
 | | 2.26 A resolution structure of MERS 3CL protease in complex with inhibitor 7i | | Descriptor: | 4,4-difluorocyclohexyl [(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6V5M
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in Complex with Succinate | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in Complex with Succinate.
To Be Published
|
|
