1OUB
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V100A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
1OUH
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V74A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
1OUF
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V130A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
3IZ2
 
 | | C-alpha model fitted into the EM structure of Cx26M34Adel2-7 | | Descriptor: | Gap junction beta-2 protein | | Authors: | Oshima, A, Tani, K, Toloue, M.M, Hiroaki, Y, Smock, A, Inukai, S, Cone, A, Nicholson, B.J, Sosinsky, G.E, Fujiyoshi, Y. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (10 Å) | | Cite: | Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels.
J.Mol.Biol., 405, 2011
|
|
3IYZ
 
 | | Structure of Aquaporin-4 S180D mutant at 10.0 A resolution from electron micrograph | | Descriptor: | Aquaporin-4 | | Authors: | Mitsuma, T, Tani, K, Hiroaki, Y, Kamegawa, A, Suzuki, H, Hibino, H, Kurachi, Y, Fujiyoshi, Y. | | Deposit date: | 2010-07-24 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (10 Å) | | Cite: | Influence of the cytoplasmic domains of aquaporin-4 on water conduction and array formation.
J.Mol.Biol., 402, 2010
|
|
5GLH
 
 | | Human endothelin receptor type-B in complex with ET-1 | | Descriptor: | Endothelin Receptor Subtype-B, Peptide from Endothelin-1 | | Authors: | Shihoya, W, Nishizawa, T, Okuta, A, Tani, K, Fujiyoshi, Y, Dohmae, N, Nureki, O, Doi, T. | | Deposit date: | 2016-07-11 | | Release date: | 2016-09-07 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation mechanism of endothelin ETB receptor by endothelin-1.
Nature, 537, 2016
|
|
5H1R
 
 | |
5GLI
 
 | | Human endothelin receptor type-B in the ligand-free form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Endothelin Receptor Subtype-B, OLEIC ACID, ... | | Authors: | Shihoya, W, Nishizawa, T, Okuta, A, Tani, K, Fujiyoshi, Y, Dohmae, N, Nureki, O, Doi, T. | | Deposit date: | 2016-07-11 | | Release date: | 2016-09-07 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Activation mechanism of endothelin ETB receptor by endothelin-1.
Nature, 537, 2016
|
|
5H1Q
 
 | |
4XRE
 
 | | Crystal structure of Gnk2 complexed with mannose | | Descriptor: | Antifungal protein ginkbilobin-2, alpha-D-mannopyranose | | Authors: | Miyakawa, T, Hatano, K, Miyauchi, Y, Suwa, Y, Sawano, Y, Tanokura, M. | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity.
Plant Physiol., 166, 2014
|
|
2D57
 
 | | Double layered 2D crystal structure of AQUAPORIN-4 (AQP4M23) at 3.2 a resolution by electron crystallography | | Descriptor: | Aquaporin-4 | | Authors: | Hiroaki, Y, Tani, K, Kamegawa, A, Gyobu, N, Nishikawa, K, Suzuki, H, Walz, T, Sasaki, S, Mitsuoka, K, Kimura, K, Mizoguchi, A, Fujiyoshi, Y. | | Deposit date: | 2005-10-29 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.2 Å) | | Cite: | Implications of the Aquaporin-4 Structure on Array Formation and Cell Adhesion
J.Mol.Biol., 355, 2005
|
|
5IAU
 
 | |
4Y6I
 
 | |
5INX
 
 | |
5IMH
 
 | |
5INR
 
 | |
5IB9
 
 | |
1YAO
 
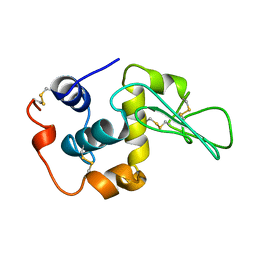 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
5IMG
 
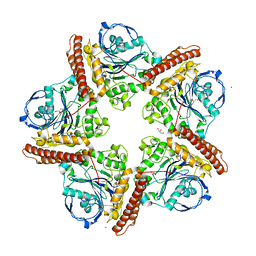 | |
1YAM
 
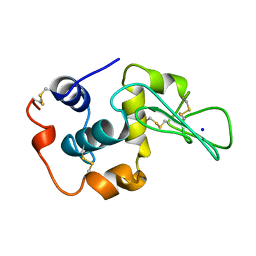 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1YAN
 
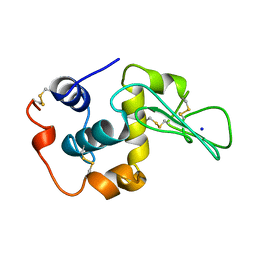 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1YAQ
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1YAP
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
4Y65
 
 | |
5B4N
 
 | | Structure analysis of function associated loop mutant of substrate recognition domain of Fbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2 | | Authors: | Nishio, K, Yoshida, Y, Tanaka, K, Mizushima, T. | | Deposit date: | 2016-04-06 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of a function-associated loop mutant of the substrate-recognition domain of Fbs1 ubiquitin ligase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
