4D1C
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH bromovinylhydantoin bound. | | Descriptor: | (5Z)-5-[(3-bromophenyl)methylidene]imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
1HJX
 
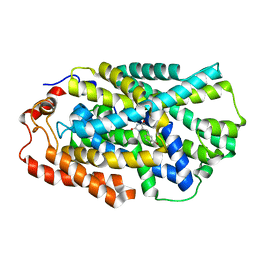
 | | Ligand-induced signalling and conformational change of the 39 kD glycoprotein from human articular chondrocytes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE-3 LIKE PROTEIN 1, GLYCEROL, ... | | Authors: | Houston, D.R, Recklies, A.D, Krupa, J.C, Van Aalten, D.M.F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Ligand-Induced Conformational Change of the 39-kDa Glycoprotein from Human Articular Chondrocytes
J.Biol.Chem., 278, 2003
|
|
3ZUX
 
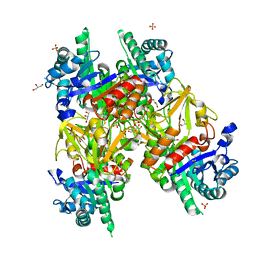
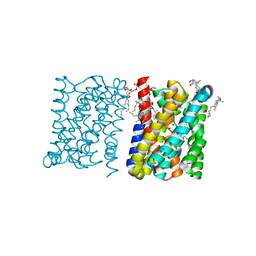 | | Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, MERCURY (II) ION, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Hu, N.-J, Iwata, S, Cameron, A.D, Drew, D. | | Deposit date: | 2011-07-21 | | Release date: | 2011-10-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Bacterial Homologue of the Bile Acid Sodium Symporter Asbt.
Nature, 478, 2011
|
|
3ZUY
 
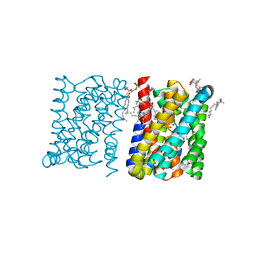 | | Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Hu, N.-J, Iwata, S, Cameron, A.D, Drew, D. | | Deposit date: | 2011-07-21 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Bacterial Homologue of the Bile Acid Sodium Symporter Asbt.
Nature, 478, 2011
|
|
4D1D
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER with the inhibitor 5-(2-naphthylmethyl)-L-hydantoin. | | Descriptor: | 5-(2-NAPHTHYLMETHYL)-D-HYDANTOIN, 5-(2-NAPHTHYLMETHYL)-L-HYDANTOIN, HYDANTOIN TRANSPORT PROTEIN, ... | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1A
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH INDOLYLMETHYL-HYDANTOIN | | Descriptor: | (5S)-5-(1H-indol-3-ylmethyl)imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
1J5B
 
 | | Solution structure of a hydrophobic analogue of the winter flounder antifreeze protein | | Descriptor: | Antifreeze protein type 1 analogue | | Authors: | Liepinsh, E, Otting, G, Harding, M.M, Ward, L.G, Mackay, J.P, Haymet, A.D. | | Deposit date: | 2002-03-22 | | Release date: | 2002-03-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a hydrophobic analogue of the winter flounder antifreeze protein.
Eur.J.Biochem., 269, 2002
|
|
4D1B
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH BENZYL-HYDANTOIN | | Descriptor: | (5S)-5-benzylimidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Brueckner, F, Geng, T, Weyand, S, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4ATV
 
 | | STRUCTURE OF A TRIPLE MUTANT OF THE NHAA DIMER, CRYSTALLISED AT LOW PH | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, NA(+)/H(+) ANTIPORTER NHAA, SULFATE ION | | Authors: | Drew, D, Lee, C, Iwata, S, Cameron, A.D. | | Deposit date: | 2012-05-10 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights.
J. Gen. Physiol., 144, 2014
|
|
4B20
 
 | | Structural basis of DNA loop recognition by Endonuclease V | | Descriptor: | 5'-D(*AP*TP*CP*TP*TP*GP*TP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*AP*CP*AP*GP)-3', ENDONUCLEASE V, ... | | Authors: | Rosnes, I, Rowe, A.D, Forstrom, R.J, Alseth, I, Bjoras, M, Dalhus, B. | | Deposit date: | 2012-07-12 | | Release date: | 2013-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of DNA Loop Recognition by Endonuclease V.
Structure, 21, 2013
|
|
4DEL
 
 | | Active site loop dynamics of a class IIa fructose 1,6-bisphosphate aldolase from M. tuberculosis | | Descriptor: | ACETATE ION, Fructose-bisphosphate aldolase, HEXAETHYLENE GLYCOL, ... | | Authors: | Pegan, S.D, Mesecar, A.D. | | Deposit date: | 2012-01-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Active site loop dynamics of a class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis.
Biochemistry, 52, 2013
|
|
3K0X
 
 | | Crystal structure of telomere capping protein Ten1 from Saccharomyces pombe | | Descriptor: | IODIDE ION, Protein Ten1 | | Authors: | Gelinas, A.D, Reyes, F.E, Batey, R.T, Wuttke, D.S. | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3KJT
 
 | |
3MKJ
 
 | | Methionine gamma-lyase from Citrobacter freundii with pyridoximine-5'-phosphate | | Descriptor: | Methionine gamma-lyase, [5-hydroxy-4-(iminomethyl)-6-methyl-pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Revtovich, S.V, Nikulin, A.D, Morozova, E.A, Demidkina, T.V. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Exploring methionine gamma-lyase structure-function relationship via microspectrophotometry and X-ray crystallography
Biochim.Biophys.Acta, 1814, 2011
|
|
3M20
 
 | | Crystal structure of DmpI from Archaeoglobus fulgidus determined to 2.37 Angstroms resolution | | Descriptor: | 4-oxalocrotonate tautomerase, putative | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J, Dasgupta, R, Kern, A.D. | | Deposit date: | 2010-03-06 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Kinetic and structural characterization of DmpI from Helicobacter pylori and Archaeoglobus fulgidus, two 4-oxalocrotonate tautomerase family members.
Bioorg.Chem., 38, 2010
|
|
3M21
 
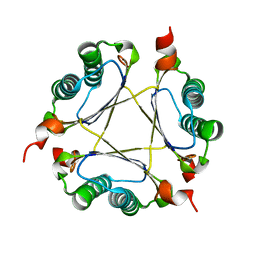 | | Crystal structure of DmpI from Helicobacter pylori Determined to 1.9 Angstroms resolution | | Descriptor: | Probable tautomerase HP_0924 | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J, Dasgupta, R, Kern, A.D, Czerwinski, R.M. | | Deposit date: | 2010-03-06 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural characterization of DmpI from Helicobacter pylori and Archaeoglobus fulgidus, two 4-oxalocrotonate tautomerase family members.
Bioorg.Chem., 38, 2010
|
|
3JWA
 
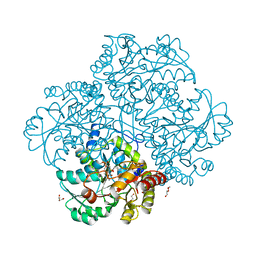 | | Crystal structure of L-methionine gamma-lyase from Citrobacter freundii with methionine phosphinate | | Descriptor: | (1-AMINO-3-METHYLSULFANYL-PROPYL)-PHOSPHINIC ACID, DI(HYDROXYETHYL)ETHER, Methionine gamma-lyase | | Authors: | Revtovish, S.V, Nikulin, A.D, Morozova, E.A, Demidkina, T.V. | | Deposit date: | 2009-09-18 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Three-dimensional structures of noncovalent complexes of Citrobacter freundii methionine gamma-lyase with substrates.
Biochemistry Mosc., 76, 2011
|
|
3JWB
 
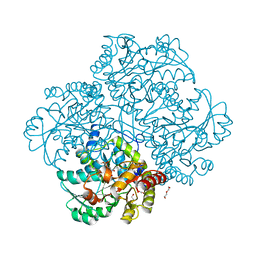 | | Crystal structure of L-methionine gamma-lyase from Citrobacter freundii with norleucine | | Descriptor: | DI(HYDROXYETHYL)ETHER, Methionine gamma-lyase, NORLEUCINE | | Authors: | Revtovish, S.V, Nikulin, A.D, Morozova, E.A, Demidkina, T.V. | | Deposit date: | 2009-09-18 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Three-dimensional structures of noncovalent complexes of Citrobacter freundii methionine gamma-lyase with substrates.
Biochemistry Mosc., 76, 2011
|
|
3JW9
 
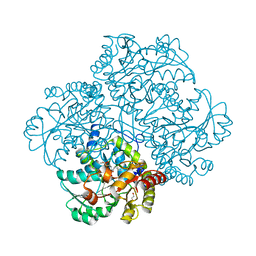 | | Crystal structure of L-methionine gamma-lyase from Citrobacter freundii with S-ethyl-cysteine | | Descriptor: | DI(HYDROXYETHYL)ETHER, Methionine gamma-lyase, S-ethyl-L-cysteine | | Authors: | Revtovish, S.V, Nikulin, A.D, Morozova, E.A, Demidkina, T.V. | | Deposit date: | 2009-09-18 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structures of noncovalent complexes of Citrobacter freundii methionine gamma-lyase with substrates.
Biochemistry Mosc., 76, 2011
|
|
3MYZ
 
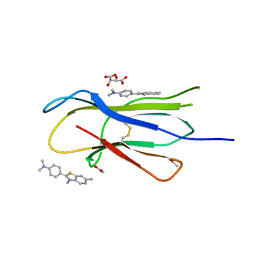 | | Protein induced photophysical changes to the amyloid indicator dye, thioflavin T | | Descriptor: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Wolfe, L.S, Calabrese, M.F, Nath, A, Blaho, D.V, Miranker, A.D, Xiong, Y. | | Deposit date: | 2010-05-11 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein-induced photophysical changes to the amyloid indicator dye thioflavin T.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MZT
 
 | | Protein-induced photophysical changes to the amyloid indicator dye, thioflavin T | | Descriptor: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, Beta-2-microglobulin, COPPER (II) ION | | Authors: | Wolfe, L.S, Calabrese, M.F, Nath, A, Blaho, D.V, Miranker, A.D, Xiong, Y. | | Deposit date: | 2010-05-13 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein-induced photophysical changes to the amyloid indicator dye thioflavin T.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MB2
 
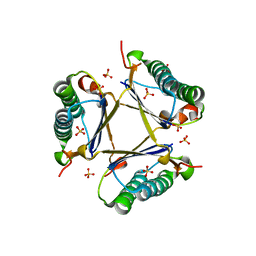 | | Kinetic and Structural Characterization of a Heterohexamer 4-Oxalocrotonate Tautomerase from Chloroflexus aurantiacus J-10-fl: Implications for Functional and Structural Diversity in the Tautomerase Superfamily | | Descriptor: | 4-oxalocrotonate tautomerase family enzyme - alpha subunit, 4-oxalocrotonate tautomerase family enzyme - beta subunit, SULFATE ION | | Authors: | Burks, E.A, Fleming, C.D, Mesecar, A.D, Whitman, C.P, Pegan, S.D. | | Deposit date: | 2010-03-24 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Kinetic and structural characterization of a heterohexamer 4-oxalocrotonate tautomerase from Chloroflexus aurantiacus J-10-fl: implications for functional and structural diversity in the tautomerase superfamily
Biochemistry, 49, 2010
|
|
3NBK
 
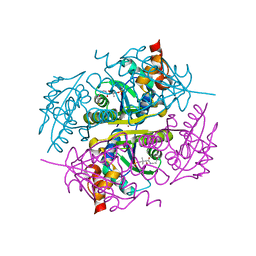 | |
3MJ5
 
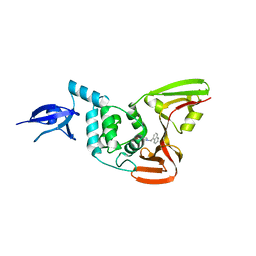 | | Severe Acute Respiratory Syndrome-Coronavirus Papain-Like Protease Inhibitors: Design, Synthesis, Protein-Ligand X-ray Structure and Biological Evaluation | | Descriptor: | N-(1,3-benzodioxol-5-ylmethyl)-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Replicase polyprotein 1a, ZINC ION | | Authors: | Mesecar, A.D, Ratia, K.M, Pegan, S.D. | | Deposit date: | 2010-04-12 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein-ligand X-ray structure and biological evaluation
J.Med.Chem., 53, 2010
|
|
3NBA
 
 | | Phosphopantetheine Adenylyltranferase from Mycobacterium tuberculosis in complex with adenosine-5'-[(alpha,beta)-methyleno]triphosphate (AMPCPP) | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, Phosphopantetheine adenylyltransferase | | Authors: | Wubben, T.J, Mesecar, A.D. | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Kinetic, thermodynamic, and structural insight into the mechanism of phosphopantetheine adenylyltransferase from Mycobacterium tuberculosis.
J.Mol.Biol., 404, 2010
|
|
