7FDK
 
 | | SARS-COV-2 Spike RBDMACSp36 binding to mACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDH
 
 | | SARS-COV-2 Spike RBDMACSp25 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDI
 
 | | SARS-COV-2 Spike RBDMACSp36 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
2BGQ
 
 | | apo aldose reductase from barley | | Descriptor: | ALDOSE REDUCTASE, SULFATE ION | | Authors: | Olsen, J.G, Pedersen, L, Christensen, C.L, Olsen, O, Henriksen, A. | | Deposit date: | 2005-01-04 | | Release date: | 2006-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Barley Aldose Reductase: Structure, Cofactor Binding, and Substrate Recognition in the Aldo/Keto Reductase 4C Family.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
7FDG
 
 | | SARS-COV-2 Spike RBDMACSp6 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
5IEU
 
 | | Crystal Structure of Mycobacterium Tuberculosis ATP-independent Proteasome Activator Tetramer | | Descriptor: | Bacterial proteasome activator | | Authors: | Bai, L, Hu, K, Wang, T, Jastrab, J.B, Darwin, K.H, Li, H. | | Deposit date: | 2016-02-25 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of the dodecameric proteasome activator PafE in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4I1W
 
 | | 2.00 Angstroms X-ray crystal structure of NAD- bound 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-11-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
2BGS
 
 | | HOLO ALDOSE REDUCTASE FROM BARLEY | | Descriptor: | ALDOSE REDUCTASE, BICARBONATE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Olsen, J.G, Pedersen, L, Christensen, C.L, Olsen, O, Henriksen, A. | | Deposit date: | 2005-01-05 | | Release date: | 2006-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Barley Aldose Reductase: Structure, Cofactor Binding, and Substrate Recognition in the Aldo/Keto Reductase 4C Family.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
5IH3
 
 | | Crystal structure of mouse CARM1 in complex with SAH at 1.8 Angstroms resolution | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with SAH at 1.8 Angstroms resolution
To Be Published
|
|
2BSM
 
 | | Novel, potent small molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design | | Descriptor: | 5-(5-CHLORO-2,4-DIHYDROXYPHENYL)-N-ETHYL-4-(4-METHOXYPHENYL)-1H-PYRAZOLE-3-CARBOXAMIDE, HEAT SHOCK PROTEIN HSP90-ALPHA | | Authors: | Dymock, B.W, Barril, X, Brough, P.A, Cansfield, J.E, Massey, A, McDonald, E, Hubbard, R.E, Surgenor, A, Roughley, S.D, Webb, P, Workman, P, Wright, L, Drysdale, M.J. | | Deposit date: | 2005-05-23 | | Release date: | 2005-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Novel, potent small-molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design.
J. Med. Chem., 48, 2005
|
|
7SUA
 
 | | Crystal Structure of the Hypothetical Protein (ACX60_00475) from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DUF4175 domain-containing protein | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Hypothetical Protein (ACX60_00475) from Acinetobacter baumannii
To be Published
|
|
2BL8
 
 | | 1.6 Angstrom crystal structure of EntA-im: a bacterial immunity protein conferring immunity to the antimicrobial activity of the pediocin-like bacteriocin, enterocin A | | Descriptor: | CITRATE ANION, ENTEROCINE A IMMUNITY PROTEIN | | Authors: | Johnsen, L, Dalhus, B, Leiros, I, Nissen-Meyer, J. | | Deposit date: | 2005-03-02 | | Release date: | 2005-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6-A Crystal Structure of Enta-Im: A Bacterial Immunity Protein Conferring Immunity to the Antimicrobial Activity of the Pediocin-Like Bacteriocin Enterocin A
J.Biol.Chem., 280, 2005
|
|
2BS1
 
 | | MS2 (N87AE89K mutant) - Qbeta RNA hairpin complex | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *UP*AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-05-13 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
2BT0
 
 | | Novel, potent small molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design | | Descriptor: | 4-[4-(2,3-DIHYDRO-1,4-BENZODIOXIN-6-YL)-3-METHYL-1H-PYRAZOL-5-YL]-6-ETHYLBENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP90-ALPHA | | Authors: | Dymock, B.W, Barril, X, Brough, P.A, Cansfield, J.E, Massey, A, McDonald, E, Hubbard, R.E, Surgenor, A, Roughley, S.D, Webb, P, Workman, P, Wright, L, Drysdale, M.J. | | Deposit date: | 2005-05-24 | | Release date: | 2005-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel, potent small-molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design.
J. Med. Chem., 48, 2005
|
|
7T1M
 
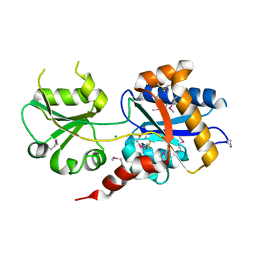 | |
2C0K
 
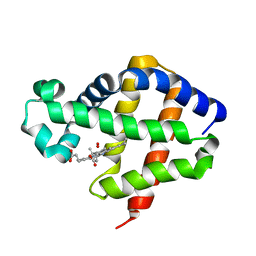 | | The structure of hemoglobin from the botfly Gasterophilus intestinalis | | Descriptor: | HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Hoogewijs, D, Ascenzi, P, Moens, L, Bolognesi, M. | | Deposit date: | 2005-09-05 | | Release date: | 2005-11-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Modulation of Oxygen Binding to Insect Hemoglobins: The Structure of Hemoglobin from the Botfly Gasterophilus Intestinalis.
Protein Sci., 14, 2005
|
|
7T1S
 
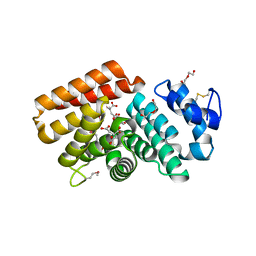 | |
6UZ3
 
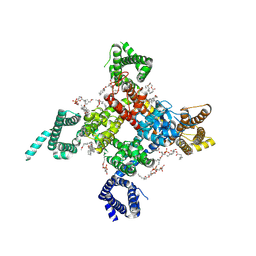 | | Cardiac sodium channel | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Shi, H, Tonggu, L, Lenaeus, M.J, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Cardiac Sodium Channel.
Cell, 180, 2020
|
|
2BNE
 
 | | The structure of E. coli UMP kinase in complex with UMP | | Descriptor: | GLYCEROL, URIDINE-5'-MONOPHOSPHATE, URIDYLATE KINASE | | Authors: | Briozzo, P, Evrin, C, Meyer, P, Assairi, L, Joly, N, Barzu, O, Gilles, A.M. | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Escherichia Coli Ump Kinase Differs from that of Other Nucleoside Monophosphate Kinases and Sheds New Light on Enzyme Regulation.
J.Biol.Chem., 280, 2005
|
|
6UYQ
 
 | | Crystal structure of K45-acetylated SUMO1 in complex with PML-SIM | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6UYR
 
 | | Crystal structure of K46-acetylated SUMO1 in complex with PML-SIM | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
4I26
 
 | | 2.20 Angstroms X-ray crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | 1,2-ETHANEDIOL, 2-aminomuconate 6-semialdehyde dehydrogenase, SODIUM ION | | Authors: | Davis, I, Huo, L, Chen, L, Liu, A. | | Deposit date: | 2012-11-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
6UYY
 
 | | Crystal structure of K39-acetylated SUMO1 in complex with phosphorylated DAXX | | Descriptor: | Small ubiquitin-related modifier 1, phosphorylated DAXX | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6VFP
 
 | | Crystal structure of human protocadherin 1 EC1-EC4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin-1, ... | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
5ISB
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0435 | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, 5'-{[(3S)-3-amino-3-carboxypropyl][(1H-imidazol-4-yl)methyl]amino}-5'-deoxyadenosine, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0435
To Be Published
|
|
