6CG7
 
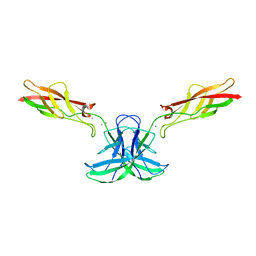 | | mouse cadherin-22 EC1-2 adhesive fragment | | Descriptor: | CALCIUM ION, Cadherin-22 | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2018-02-19 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior.
Cell Rep, 23, 2018
|
|
6CGS
 
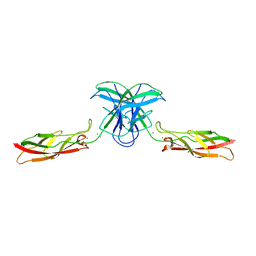 | | mouse cadherin-7 EC1-2 adhesive fragment | | Descriptor: | CALCIUM ION, Cadherin-7, GLYCEROL | | Authors: | Brasch, J, Harrison, O.J, Kaczynska, A, Shapiro, L. | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior.
Cell Rep, 23, 2018
|
|
4X3H
 
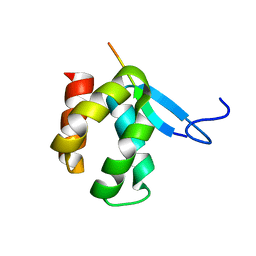 | | CRYSTAL STRUCTURE OF ARC N-LOBE COMPLEXED WITH STARGAZIN PEPTIDE | | Descriptor: | Activity-regulated cytoskeleton-associated protein, VOLTAGE-DEPENDENT CALCIUM CHANNEL GAMMA-2 SUBUNIT | | Authors: | zhang, W, ward, m, leahy, d, worley, p. | | Deposit date: | 2014-11-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
4X3I
 
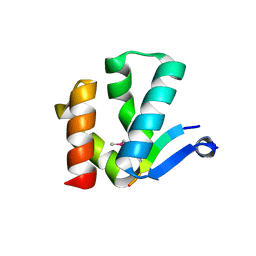 | | The crystal structure of Arc N-lobe complexed with CAMK2A fragment | | Descriptor: | Activity-regulated cytoskeleton-associated protein, CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE TYPE II SUBUNIT ALPHA | | Authors: | Zhang, W, Ward, M, Leahy, D, Worley, P. | | Deposit date: | 2014-11-30 | | Release date: | 2015-06-03 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
2Q13
 
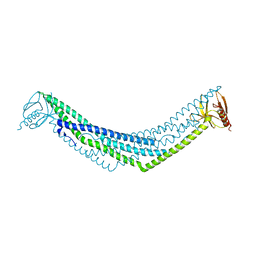 | | Crystal structure of BAR-PH domain of APPL1 | | Descriptor: | DCC-interacting protein 13 alpha | | Authors: | Zhu, G, Zhang, X.C. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5.
Embo J., 26, 2007
|
|
5CF8
 
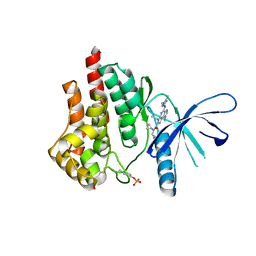 | | CRYSTAL STRUCTURE OF JANUS KINASE 2 IN COMPLEX WITH N,N-DICYCLOPROPYL-10-ETHYL-7-[(3-METHOXYPROPYL)AMINO] -3-METHYL-3,5,8,10-TETRAAZATRICYCLO[7.3.0.0,6] DODECA-1(9),2(6),4,7,11-PENTAENE-11-CARBOXAMIDE | | Descriptor: | N,N-dicyclopropyl-4-[(1,5-dimethyl-1H-pyrazol-3-yl)amino]-6-ethyl-1-methyl-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Sack, J.S. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Highly Selective JAK2 Inhibitor, BMS-911543, for the Treatment of Myeloproliferative Neoplasms.
Acs Med.Chem.Lett., 6, 2015
|
|
1NSC
 
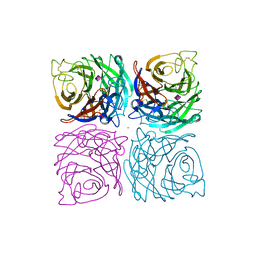 | | INFLUENZA B VIRUS NEURAMINIDASE CAN SYNTHESIZE ITS OWN INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Burmeister, W.P, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 1993-05-24 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influenza B virus neuraminidase can synthesize its own inhibitor.
Structure, 1, 1993
|
|
2Q12
 
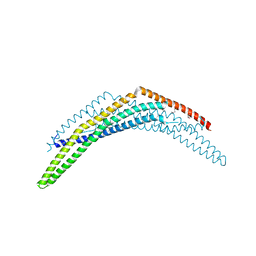 | | Crystal Structure of BAR domain of APPL1 | | Descriptor: | DCC-interacting protein 13 alpha | | Authors: | Zhang, X.C, Zhu, G. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5.
Embo J., 26, 2007
|
|
1NSD
 
 | | INFLUENZA B VIRUS NEURAMINIDASE CAN SYNTHESIZE ITS OWN INHIBITOR | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Burmeister, W.P, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 1993-05-24 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influenza B virus neuraminidase can synthesize its own inhibitor.
Structure, 1, 1993
|
|
2VOZ
 
 | | Apo FutA2 from Synechocystis PCC6803 | | Descriptor: | PERIPLASMIC IRON-BINDING PROTEIN, SULFATE ION | | Authors: | Badarau, A, Firbank, S.J, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-02-25 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Futa2 is a Ferric Binding Protein from Synechocystis Pcc 6803.
J.Biol.Chem., 283, 2008
|
|
6IHL
 
 | | Crystal structure of bacterial serine phosphatase | | Descriptor: | MAGNESIUM ION, Phosphorylated protein phosphatase | | Authors: | Yang, C.-G, yang, T. | | Deposit date: | 2018-09-30 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Structural Insight into the Mechanism of Staphylococcus aureus Stp1 Phosphatase.
Acs Infect Dis., 5, 2019
|
|
6IHW
 
 | |
6IHR
 
 | |
6IHS
 
 | |
6IHT
 
 | |
1NSB
 
 | |
6IHU
 
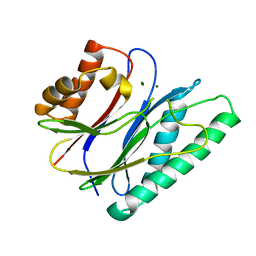 | |
6IHV
 
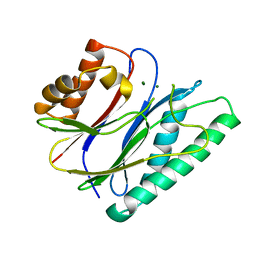 | |
3TKJ
 
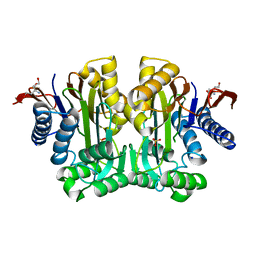 | | Crystal Structure of Human Asparaginase-like Protein 1 Thr168Ala | | Descriptor: | L-asparaginase, SODIUM ION, SULFATE ION, ... | | Authors: | Li, W.Z, Yogesha, S.D, Liu, J, Zhang, Y. | | Deposit date: | 2011-08-26 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uncoupling Intramolecular Processing and Substrate Hydrolysis in the N-Terminal Nucleophile Hydrolase hASRGL1 by Circular Permutation.
Acs Chem.Biol., 7, 2012
|
|
7K1M
 
 | |
8WHZ
 
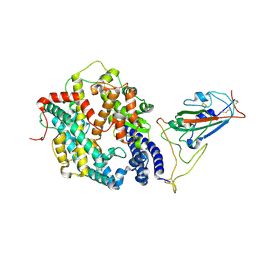 | | BA.2.86 RBD in complex with hACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHS
 
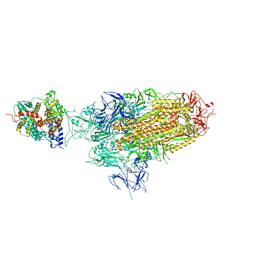 | | Spike Trimer of BA.2.86 in complex with one hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHV
 
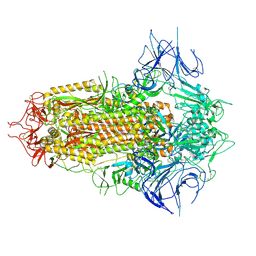 | | Spike Trimer of BA.2.86 with three RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHW
 
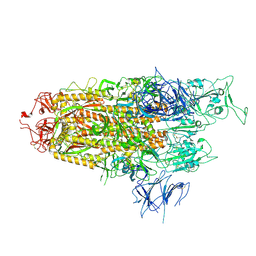 | | Spike Trimer of BA.2.86 with single RBD up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHU
 
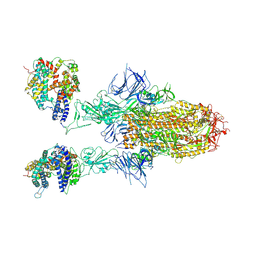 | | Spike Trimer of BA.2.86 in complex with two hACE2s | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
