4G99
 
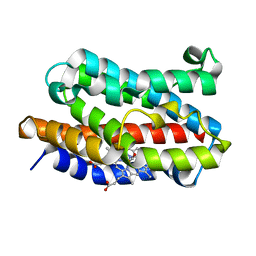 | | Rat Heme Oxygenase-1 in complex with Heme and CO at 100 K after warming to 160 K | | Descriptor: | CARBON MONOXIDE, FORMIC ACID, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Moffat, K, Noguchi, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discrimination between CO and O(2) in heme oxygenase: comparison of static structures and dynamic conformation changes following CO photolysis.
Biochemistry, 51, 2012
|
|
1ULX
 
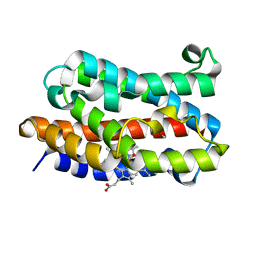 | | Partially photolyzed structure of CO-bound heme-heme oxygenase complex | | Descriptor: | CARBON MONOXIDE, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2003-09-16 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CO-trapping site in heme oxygenase revealed by photolysis of its co-bound heme complex: mechanism of escaping from product inhibition
J.Mol.Biol., 341, 2004
|
|
1VGI
 
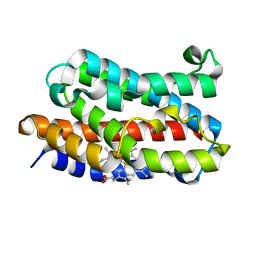 | | Crystal structure of xenon bound rat heme-heme oxygenase-1 complex | | Descriptor: | FORMIC ACID, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sugishima, M, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2004-04-26 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CO-trapping site in heme oxygenase revealed by photolysis of its co-bound heme complex: mechanism of escaping from product inhibition
J.Mol.Biol., 341, 2004
|
|
3WKT
 
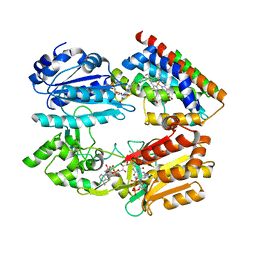 | | Complex structure of an open form of NADPH-cytochrome P450 reductase and heme oxygenase-1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Sato, H, Higashimoto, Y, Harada, J, Wada, K, Fukuyama, K, Noguchi, M. | | Deposit date: | 2013-10-31 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Structural basis for the electron transfer from an open form of NADPH-cytochrome P450 oxidoreductase to heme oxygenase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2ZVU
 
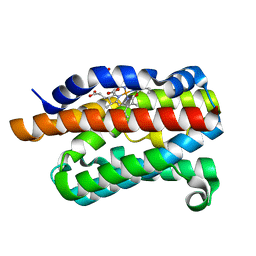 | | Crystal structure of rat heme oxygenase-1 in complex with ferrous verdoheme | | Descriptor: | 5-OXA-PROTOPORPHYRIN IX CONTAINING FE, FORMIC ACID, Heme oxygenase 1 | | Authors: | Sato, H, Sugishima, M, Fukuyama, K, Noguchi, M. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of rat haem oxygenase-1 in complex with ferrous verdohaem: presence of a hydrogen-bond network on the distal side
Biochem.J., 419, 2009
|
|
2DY5
 
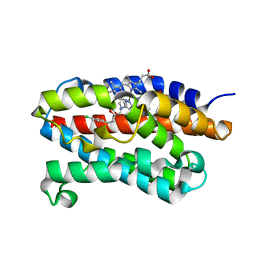 | | Crystal structure of rat heme oxygenase-1 in complex with heme and 2-[2-(4-chlorophenyl)ethyl]-2-[(1H-imidazol-1-yl)methyl]-1,3-dioxolane | | Descriptor: | 1-({2-[2-(4-CHLOROPHENYL)ETHYL]-1,3-DIOXOLAN-2-YL}METHYL)-1H-IMIDAZOLE, CHLORIDE ION, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Takahashi, H, Fukuyama, K. | | Deposit date: | 2006-09-06 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray crystallographic and biochemical characterization of the inhibitory action of an imidazole-dioxolane compound on heme oxygenase
Biochemistry, 46, 2007
|
|
2E7E
 
 | |
3AGC
 
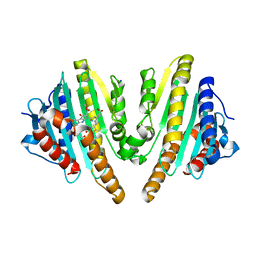 | | F218V mutant of the substrate-bound red chlorophyll catabolite reductase from Arabidopsis thaliana | | Descriptor: | 3-{(2Z,3S,4S)-5-[(Z)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-2-[(5R)-2-[(3-ethyl-5-formyl-4-methyl-1H-pyrrol-2-yl)methyl]-5-(methoxycarbonyl)-3-methyl-4-oxo-4,5-dihydrocyclopenta[b]pyrrol-6(1H)-ylidene]-4-methyl-3,4-dihydro-2H-pyrrol-3-yl}propanoic acid, Red chlorophyll catabolite reductase, chloroplastic, ... | | Authors: | Sugishima, M, Fukuyama, K. | | Deposit date: | 2010-03-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the substrate-bound forms of red chlorophyll catabolite reductase: implications for site-specific and stereospecific reaction
J.Mol.Biol., 402, 2010
|
|
3AGA
 
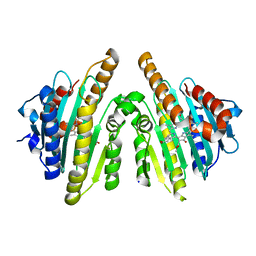 | | Crystal structure of RCC-bound red chlorophyll catabolite reductase from Arabidopsis thaliana | | Descriptor: | 3-{(2Z,3S,4S)-5-[(Z)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-2-[(5R)-2-[(3-ethyl-5-formyl-4-methyl-1H-pyrrol-2-yl)methyl]-5-(methoxycarbonyl)-3-methyl-4-oxo-4,5-dihydrocyclopenta[b]pyrrol-6(1H)-ylidene]-4-methyl-3,4-dihydro-2H-pyrrol-3-yl}propanoic acid, Red chlorophyll catabolite reductase, chloroplastic, ... | | Authors: | Sugishima, M, Fukuyama, K. | | Deposit date: | 2010-03-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the substrate-bound forms of red chlorophyll catabolite reductase: implications for site-specific and stereospecific reaction
J.Mol.Biol., 402, 2010
|
|
3AGB
 
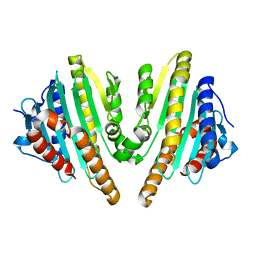 | |
1J02
 
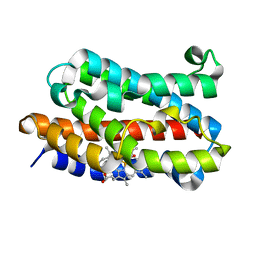 | | Crystal Structure of Rat Heme Oxygenase-1-Heme Bound to NO | | Descriptor: | HEME OXYGENASE 1, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Fukuyama, K. | | Deposit date: | 2002-10-28 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
2ZXL
 
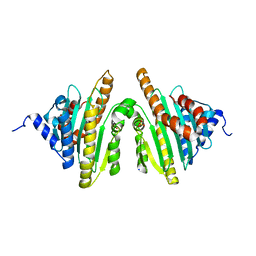 | | Crystal structure of red chlorophyll catabolite reductase from Arabidopsis thaliana | | Descriptor: | Red chlorophyll catabolite reductase, chloroplastic, SODIUM ION, ... | | Authors: | Sugishima, M, Kitamori, Y, Fukuyama, K. | | Deposit date: | 2008-12-29 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of red chlorophyll catabolite reductase: enlargement of the ferredoxin-dependent bilin reductase family
J.Mol.Biol., 389, 2009
|
|
2ZXK
 
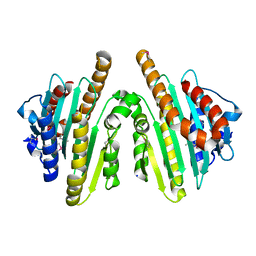 | |
3UG4
 
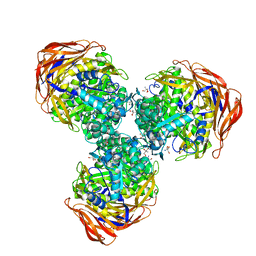 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
3UG5
 
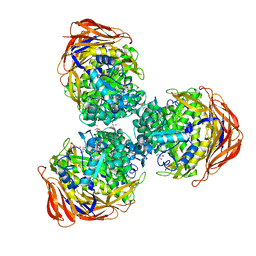 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, beta-D-xylopyranose | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
3UG3
 
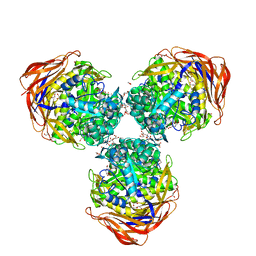 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, ... | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
