4DLB
 
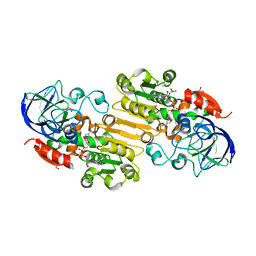 | | Structure of S-nitrosoglutathione reductase from tomato (Solanum lycopersicum) crystallized in presence of NADH and glutathione | | Descriptor: | Alcohol dehydrogenase class III, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kopecny, D, Tylichova, M, Briozzo, P. | | Deposit date: | 2012-02-06 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional characterization of a plant S-nitrosoglutathione reductase from Solanum lycopersicum.
Biochimie, 95, 2013
|
|
3IWK
 
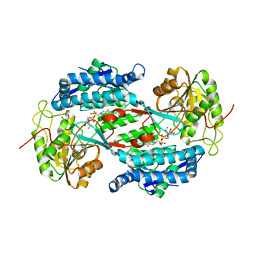 | | Crystal structure of aminoaldehyde dehydrogenase 1 from Pisum sativum (PsAMADH1) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Aminoaldehyde dehydrogenase, GLYCEROL, ... | | Authors: | Kopecny, D, Morera, S, Briozzo, P. | | Deposit date: | 2009-09-02 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of plant aminoaldehyde dehydrogenase from Pisum sativum with a broad specificity for natural and synthetic aminoaldehydes.
J.Mol.Biol., 396, 2010
|
|
3BW7
 
 | |
3C0P
 
 | |
3DQ0
 
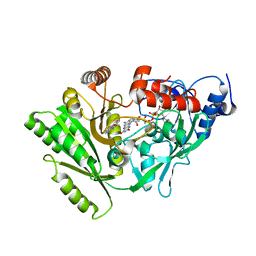 | | Maize cytokinin oxidase/dehydrogenase complexed with N6-(3-methoxy-phenyl)adenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, ... | | Authors: | Kopecny, D, Briozzo, P. | | Deposit date: | 2008-07-09 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and biological activity of novel purine-derived inhibitor of cytokinin oxidase/dehydrogenase and its potential use for in vivo studies
To be Published
|
|
4DLA
 
 | |
4I8P
 
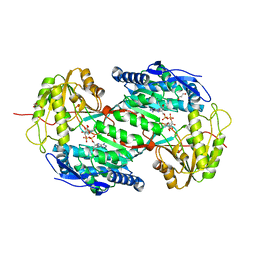 | | Crystal structure of aminoaldehyde dehydrogenase 1a from Zea mays (ZmAMADH1a) | | Descriptor: | 1,2-ETHANEDIOL, Aminoaldehyde dehydrogenase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
4I8Q
 
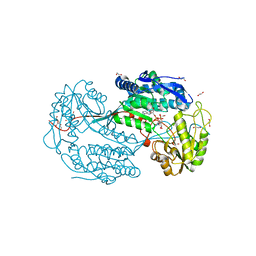 | | Structure of the aminoaldehyde dehydrogenase 1 E260A mutant from Solanum lycopersicum (SlAMADH1-E260A) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
4I9B
 
 | | Structure of aminoaldehyde dehydrogenase 1 from Solanum lycopersium (SlAMADH1) with a thiohemiacetal intermediate | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-05 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
4MLA
 
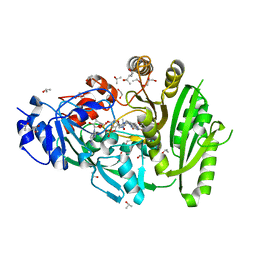 | | Structure of maize cytokinin oxidase/dehydrogenase 2 (ZmCKO2) | | Descriptor: | 1,2-ETHANEDIOL, Cytokinin oxidase 2, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Morera, S, Kopecny, D, Briozzo, P, Koncitikova, R. | | Deposit date: | 2013-09-06 | | Release date: | 2015-03-11 | | Last modified: | 2016-03-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
4ML8
 
 | | Structure of maize cytokinin oxidase/dehydrogenase 2 (ZmCKO2) | | Descriptor: | Cytokinin oxidase 2, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Morera, S, Kopecny, D, Briozzo, P, Koncitikova, R. | | Deposit date: | 2013-09-06 | | Release date: | 2015-03-11 | | Last modified: | 2016-03-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
7ZHC
 
 | | Moss spermine/spermidine acetyl transferase (PpSSAT) in complex with AcetylCoA and polyethylen glycol | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, N-acetyltransferase domain-containing protein, ... | | Authors: | Morera, S, Kopecny, D, Vigouroux, A. | | Deposit date: | 2022-04-06 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Biochemical and structural basis of polyamine, lysine and ornithine acetylation catalyzed by spermine/spermidine N-acetyl transferase in moss and maize.
Plant J., 114, 2023
|
|
7ZKT
 
 | | Moss spermine/spermidine acetyl transferase (PpSSAT) in complex with CoA and lysine | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, LYSINE, ... | | Authors: | Morera, S, Kopecny, D, Vigouroux, A, Briozzo, P. | | Deposit date: | 2022-04-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Biochemical and structural basis of polyamine, lysine and ornithine acetylation catalyzed by spermine/spermidine N-acetyl transferase in moss and maize.
Plant J., 114, 2023
|
|
8OF3
 
 | | Structure of the apoform of ALDEHYDE DEHYDROGENASE 5F1 (ALDH5F1) from the moss Physcomitrium patens | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Morera, S, Kopecny, D, Vigouroux, A. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.348 Å) | | Cite: | A study on abiotic stress responses of aldehyde dehydrogenase (ALDH) superfamilies in moss and barley focused on members linked to the GABA shunt pathway
To Be Published
|
|
8OFM
 
 | |
8OF1
 
 | | Structure of ALDH5F1 from moss Physcomitrium patens in complex with NAD+ in the contracted conformation | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Kopecny, D, Vigouroux, A. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A study on abiotic stress responses of aldehyde dehydrogenase (ALDH) superfamilies in moss and barley focused on members linked to the GABA shunt pathway
To Be Published
|
|
6ZK1
 
 | | Plant nucleoside hydrolase - ZmNRh2b enzyme | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK2
 
 | | Plant nucleoside hydrolase - ZmNRh2b in complex with forodesine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK3
 
 | | Plant nucleoside hydrolase - ZmNRh2b in complex with ribose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK4
 
 | | Plant nucleoside hydrolase - ZmNRh2b with a bound adenine | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, CALCIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK5
 
 | | Plant nucleoside hydrolase - ZmNRh3 enzyme in complex with forodesine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6D97
 
 | |
5EGW
 
 | |
5EF4
 
 | | 2.05 A crystal structure of the Amb a 11 cysteine protease, a major ragweed pollen allergen, in its proform | | Descriptor: | Cysteine protease, GLYCEROL | | Authors: | Briozzo, P, Kopecny, D, Savko, M. | | Deposit date: | 2015-10-23 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Functional Characterization of the Major Allergen Amb a 11 from Short Ragweed Pollen.
J.Biol.Chem., 291, 2016
|
|
4KPN
 
 | | Plant nucleoside hydrolase - PpNRh1 enzyme | | Descriptor: | CALCIUM ION, Nucleoside N-ribohydrolase 1 | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2013-05-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structure and Function of Nucleoside Hydrolases from Physcomitrella patens and Maize Catalyzing the Hydrolysis of Purine, Pyrimidine, and Cytokinin Ribosides.
Plant Physiol., 163, 2013
|
|
