1V4Q
 
 | |
3VMF
 
 | | Archaeal protein | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kobayashi, K, Saito, K, Ishitani, R, Ito, K, Nureki, O. | | Deposit date: | 2011-12-12 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for translation termination by archaeal RF1 and GTP-bound EF1alpha complex
Nucleic Acids Res., 40, 2012
|
|
3WXM
 
 | | Crystal structure of archaeal Pelota and GTP-bound EF1 alpha complex | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kobayashi, K, Ishitani, R, Nureki, O. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1 alpha complex
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3WTR
 
 | | Crystal structure of E. coli YfcM bound to Co(II) | | Descriptor: | COBALT (II) ION, Uncharacterized protein | | Authors: | Kobayashi, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2014-04-19 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The non-canonical hydroxylase structure of YfcM reveals a metal ion-coordination motif required for EF-P hydroxylation
Nucleic Acids Res., 42, 2014
|
|
3AGK
 
 | | Crystal structure of archaeal translation termination factor, aRF1 | | Descriptor: | Peptide chain release factor subunit 1 | | Authors: | Kobayashi, K, Kikuno, I, Ishitani, R, Ito, K, Nureki, O. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Omnipotent role of archaeal elongation factor 1 alpha (EF1{alpha}) in translational elongation and termination, and quality control of protein synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7C7L
 
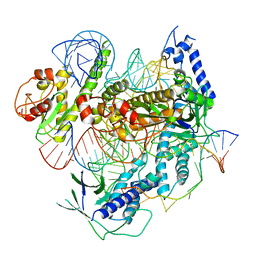 | | Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex | | Descriptor: | CRISPR-associated protein Cas14a.1, DNA (40-mer), ZINC ION, ... | | Authors: | Takeda, N.S, Nakagawa, R, Okazaki, S, Hirano, H, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2020-05-26 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the miniature type V-F CRISPR-Cas effector enzyme.
Mol.Cell, 81, 2021
|
|
8Z47
 
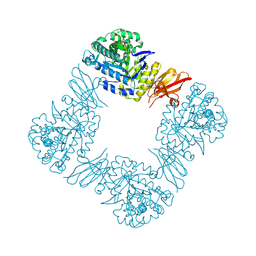 | | Beta-galactosidase from Bacteroides xylanisolvens (ligand-free) | | Descriptor: | Beta-galactosidase, alpha-D-galactopyranose | | Authors: | Nakajima, M, Motouchi, S, Kobayashi, K. | | Deposit date: | 2024-04-16 | | Release date: | 2025-01-01 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure and function of a beta-1,2-galactosidase from Bacteroides xylanisolvens, an intestinal bacterium.
Commun Biol, 8, 2025
|
|
8Z43
 
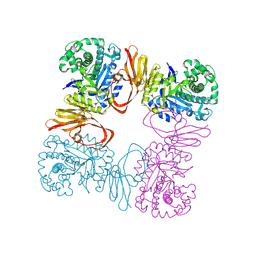 | | Beta-galactosidase from Bacteroides xylanisolvens (E350G, ligand-free) | | Descriptor: | Beta-galactosidase, GLYCEROL | | Authors: | Nakajima, M, Motouchi, S, Kobayashi, K. | | Deposit date: | 2024-04-16 | | Release date: | 2025-01-01 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure and function of a beta-1,2-galactosidase from Bacteroides xylanisolvens, an intestinal bacterium.
Commun Biol, 8, 2025
|
|
8Z48
 
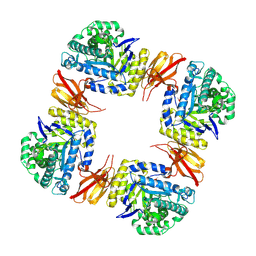 | | Beta-galactosidase from Bacteroides xylanisolvens (complex with methyl beta-galactopyranose) | | Descriptor: | Beta-galactosidase, methyl beta-D-galactopyranoside | | Authors: | Nakajima, M, Motouchi, S, Kobayashi, K. | | Deposit date: | 2024-04-16 | | Release date: | 2025-01-01 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure and function of a beta-1,2-galactosidase from Bacteroides xylanisolvens, an intestinal bacterium.
Commun Biol, 8, 2025
|
|
8Y6W
 
 | | TUG-1375 and 4-CMTB-bound human FFA2 in complex with Gi | | Descriptor: | (2R,4R)-2-(2-chlorophenyl)-3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]carbonyl-1,3-thiazolidine-4-carboxylic acid, (2~{S})-2-(4-chlorophenyl)-3-methyl-~{N}-(1,3-thiazol-2-yl)butanamide, Free fatty acid receptor 2, ... | | Authors: | Kugawa, M, Kawakami, K, Kise, R, Kobayashi, K, Kojima, A, Inoue, W, Fukuda, M, Inoue, A, Kato, H.E. | | Deposit date: | 2024-02-03 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural insights into lipid chain-length selectivity and allosteric regulation of FFA2.
Nat Commun, 16, 2025
|
|
8Y6Y
 
 | | GLPG0974-bound human FFA2 | | Descriptor: | 4-[[(2~{R})-1-(1-benzothiophen-3-ylcarbonyl)-2-methyl-azetidin-2-yl]carbonyl-[(3-chlorophenyl)methyl]amino]butanoic acid, Free fatty acid receptor 2,Soluble cytochrome b562 | | Authors: | Kugawa, M, Kawakami, K, Kise, R, Kobayashi, K, Kojima, A, Inoue, W, Fukuda, M, Inoue, A, Kato, H.E. | | Deposit date: | 2024-02-03 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural insights into lipid chain-length selectivity and allosteric regulation of FFA2.
Nat Commun, 16, 2025
|
|
7D3S
 
 | | Human SECR in complex with an engineered Gs heterotrimer | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Fukuhara, S, Kobayashi, K, Kusakizako, T, Shihoya, W, Nureki, O. | | Deposit date: | 2020-09-20 | | Release date: | 2020-11-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the human secretin receptor coupled to an engineered heterotrimeric G protein.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7YL8
 
 | |
3AQ8
 
 | | Crystal structure of truncated hemoglobin from Tetrahymena pyriformis, Q46E mutant, Fe(III) form | | Descriptor: | Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Igarashi, J, Kobayashi, K, Matsuoka, A. | | Deposit date: | 2010-10-25 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A hydrogen-bonding network formed by the B10-E7-E11 residues of a truncated hemoglobin from Tetrahymena pyriformis is critical for stability of bound oxygen and nitric oxide detoxification.
J.Biol.Inorg.Chem., 16, 2011
|
|
3AQ9
 
 | | Crystal structure of truncated hemoglobin from Tetrahymena pyriformis, Q50E mutant, Fe(III) form | | Descriptor: | Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Igarashi, J, Kobayashi, K, Matsuoka, A. | | Deposit date: | 2010-10-25 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A hydrogen-bonding network formed by the B10-E7-E11 residues of a truncated hemoglobin from Tetrahymena pyriformis is critical for stability of bound oxygen and nitric oxide detoxification.
J.Biol.Inorg.Chem., 16, 2011
|
|
3AQ6
 
 | |
3AQ5
 
 | | Crystal structure of truncated hemoglobin from Tetrahymena pyriformis, Fe(II)-O2 form | | Descriptor: | Group 1 truncated hemoglobin, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Igarashi, J, Kobayashi, K, Matsuoka, A. | | Deposit date: | 2010-10-25 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A hydrogen-bonding network formed by the B10-E7-E11 residues of a truncated hemoglobin from Tetrahymena pyriformis is critical for stability of bound oxygen and nitric oxide detoxification.
J.Biol.Inorg.Chem., 16, 2011
|
|
3AQ7
 
 | | Crystal structure of truncated hemoglobin from Tetrahymena pyriformis, Y25F mutant, Fe(III) form | | Descriptor: | Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Igarashi, J, Kobayashi, K, Matsuoka, A. | | Deposit date: | 2010-10-25 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A hydrogen-bonding network formed by the B10-E7-E11 residues of a truncated hemoglobin from Tetrahymena pyriformis is critical for stability of bound oxygen and nitric oxide detoxification.
J.Biol.Inorg.Chem., 16, 2011
|
|
5ZZN
 
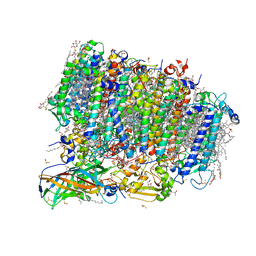 | | Crystal structure of photosystem II from an SQDG-deficient mutant of Thermosynechococcus elongatus | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Nakajima, Y, Umena, Y, Nagao, R, Endo, K, Kobayashi, K, Akita, F, Suga, M, Wada, H, Noguchi, T, Shen, J.R. | | Deposit date: | 2018-06-03 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thylakoid membrane lipid sulfoquinovosyl-diacylglycerol (SQDG) is required for full functioning of photosystem II inThermosynechococcus elongatus.
J. Biol. Chem., 293, 2018
|
|
1OMG
 
 | | NMR STUDY OF OMEGA-CONOTOXIN MVIIA | | Descriptor: | OMEGA-CONOTOXIN MVIIA | | Authors: | Kohno, T, Kim, J.-I, Kobayashi, K, Kodera, Y, Maeda, T, Sato, K. | | Deposit date: | 1995-04-26 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of the calcium channel blocker omega-conotoxin MVIIA.
Biochemistry, 34, 1995
|
|
1EHA
 
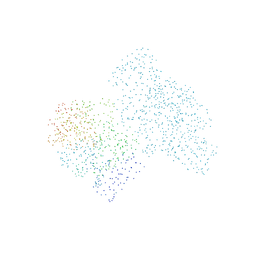 | | CRYSTAL STRUCTURE OF GLYCOSYLTREHALOSE TREHALOHYDROLASE FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | GLYCOSYLTREHALOSE TREHALOHYDROLASE | | Authors: | Feese, M.D, Kato, Y, Tamada, T, Kato, M, Komeda, T, Kobayashi, K, Kuroki, R. | | Deposit date: | 2000-02-19 | | Release date: | 2001-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of glycosyltrehalose trehalohydrolase from the hyperthermophilic archaeum Sulfolobus solfataricus.
J.Mol.Biol., 301, 2000
|
|
1EH9
 
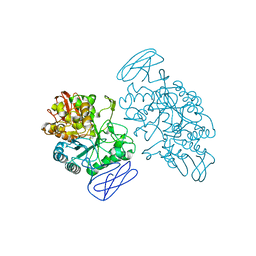 | | CRYSTAL STRUCTURE OF SULFOLOBUS SOLFATARICUS GLYCOSYLTREHALOSE TREHALOHYDROLASE | | Descriptor: | GLYCOSYLTREHALOSE TREHALOHYDROLASE | | Authors: | Feese, M.D, Kato, Y, Tamada, T, Kato, M, Komeda, T, Kobayashi, K, Kuroki, R. | | Deposit date: | 2000-02-19 | | Release date: | 2001-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of glycosyltrehalose trehalohydrolase from the hyperthermophilic archaeum Sulfolobus solfataricus.
J.Mol.Biol., 301, 2000
|
|
8WY1
 
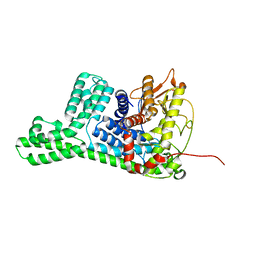 | | The structure of cyclization domain in cyclic beta-1,2-glucan synthase from Thermoanaerobacter italicus | | Descriptor: | Glycosyltransferase 36 | | Authors: | Tanaka, N, Saito, R, Kobayashi, K, Nakai, H, Kamo, S, Kuramochi, K, Taguchi, H, Nakajima, M, Masaike, T. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Functional and structural analysis of a cyclization domain in a cyclic beta-1,2-glucan synthase.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
1BM0
 
 | | CRYSTAL STRUCTURE OF HUMAN SERUM ALBUMIN | | Descriptor: | SERUM ALBUMIN | | Authors: | Sugio, S, Kashima, A, Mochizuki, S, Noda, M, Kobayashi, K. | | Deposit date: | 1998-07-28 | | Release date: | 1999-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human serum albumin at 2.5 A resolution.
Protein Eng., 12, 1999
|
|
7F8O
 
 | | Cryo-EM structure of the C-terminal deletion mutant of human PANX1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
