6K1E
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mg and GMP | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MAGNESIUM ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
6KBN
 
 | | Crystal structure of Vac8 (del 19-33) bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
6K1B
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mn and dGMP | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MANGANESE (II) ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
7X15
 
 | | Crystal structure of MIGA2 LD targeting domain | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, FORMIC ACID, Mitoguardin 2 | | Authors: | Kim, H, Lee, C. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Structural basis for mitoguardin-2 mediated lipid transport at ER-mitochondrial membrane contact sites.
Nat Commun, 13, 2022
|
|
7X14
 
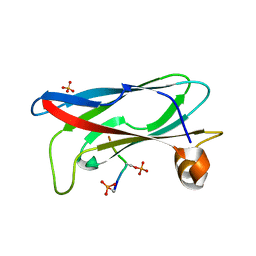 | | Crystal structure of phospho-FFAT motif of MIGA2 bound to VAPB | | Descriptor: | MIGA2 phospho FFAT motif, SULFATE ION, Vesicle-associated membrane protein-associated protein B | | Authors: | Kim, H, Lee, C. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.675 Å) | | Cite: | Structural basis for mitoguardin-2 mediated lipid transport at ER-mitochondrial membrane contact sites.
Nat Commun, 13, 2022
|
|
7YCJ
 
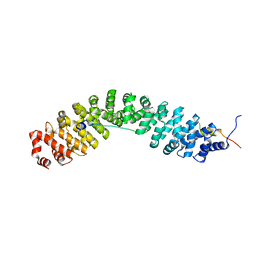 | |
4O9W
 
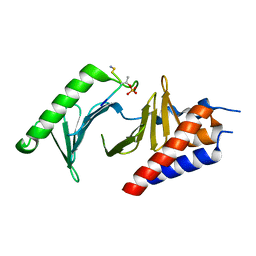 | |
8I4O
 
 | | Design of a split green fluorescent protein for sensing and tracking an beta-amyloid | | Descriptor: | Beta-amyloid, Split Green flourescent protein | | Authors: | Taegeun, Y, Jinsu, L, Jungmin, Y, Jungmin, C, Wondo, H, Song, J.J, Haksung, K. | | Deposit date: | 2023-01-20 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Engineering of a Fluorescent Protein for a Sensing of an Intrinsically Disordered Protein through Transition in the Chromophore State.
Jacs Au, 3, 2023
|
|
2LRK
 
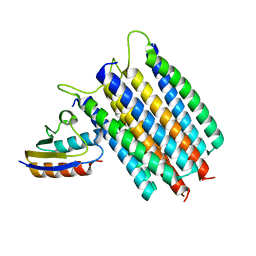 | | Solution Structures of the IIA(Chitobiose)-HPr complex of the N,N'-Diacetylchitobiose | | Descriptor: | N,N'-diacetylchitobiose-specific phosphotransferase enzyme IIA component, Phosphocarrier protein HPr | | Authors: | Cai, M, Jung, Y, Clore, M. | | Deposit date: | 2012-04-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the IIAChitobiose-HPr Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia coli Phosphotransferase System.
J.Biol.Chem., 287, 2012
|
|
