1JK4
 
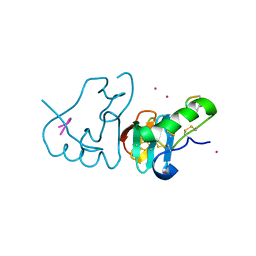 | | DES 1-6 BOVINE NEUROPHYSIN II COMPLEX WITH VASOPRESSIN | | Descriptor: | CADMIUM ION, Lys Vasopressin, NEUROPHYSIN 2 | | Authors: | Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-07-11 | | Release date: | 2003-02-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of an Unliganded Neurophysin and its Vasopressin Complex: Implications for Binding and Allosteric Mechanisms
Protein Sci., 10, 2001
|
|
1JK6
 
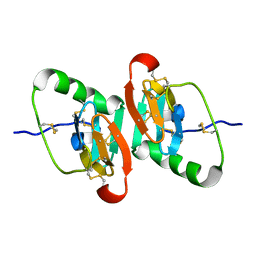 | | UNCOMPLEXED DES 1-6 BOVINE NEUROPHYSIN | | Descriptor: | NEUROPHYSIN 2 | | Authors: | Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-07-11 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of an Unliganded Neurophysin and its Vasopressin Complex: Implications for Binding and Allosteric Mechanisms
Protein Sci., 10, 2003
|
|
6B3F
 
 | |
6B38
 
 | |
6B3G
 
 | |
6B36
 
 | |
6B3C
 
 | |
6B3H
 
 | | Crystal Structure of HIV Protease complexed with N-(2-(2-((6R,9S)-2,2-dioxido-2-thia-1,7-diazabicyclo[4.3.1]decan-9-yl)ethyl)-3-fluorophenyl)-3,3-bis(4-fluorophenyl)propanamide | | Descriptor: | CHLORIDE ION, HIV-1 Protease, N-(2-{2-[(6R,9S)-2,2-dioxo-2lambda~6~-thia-1,7-diazabicyclo[4.3.1]decan-9-yl]ethyl}-3-fluorophenyl)-3,3-bis(4-fluorophenyl)propanamide | | Authors: | Su, H.P. | | Deposit date: | 2017-09-21 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Design and Synthesis of Piperazine Sulfonamide Cores Leading to Highly Potent HIV-1 Protease Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
6NM5
 
 | | F-pilus/MS2 Maturation protein complex | | Descriptor: | (2R)-2,3-dihydroxypropyl ethyl hydrogen (S)-phosphate, Maturation protein, Type IV conjugative transfer system pilin TraA | | Authors: | Meng, R, Chang, J, Zhang, J. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural basis for the adsorption of a single-stranded RNA bacteriophage.
Nat Commun, 10, 2019
|
|
7JUP
 
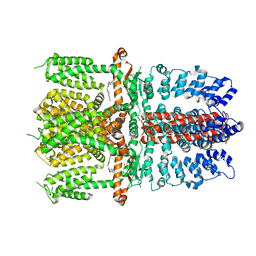 | | Structure of human TRPA1 in complex with antagonist compound 21 | | Descriptor: | 1-({3-[(3R,5R)-5-(4-fluorophenyl)oxolan-3-yl]-1,2,4-oxadiazol-5-yl}methyl)-7-methyl-1,7-dihydro-6H-purin-6-one, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L. | | Deposit date: | 2020-08-20 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Tetrahydrofuran-Based Transient Receptor Potential Ankyrin 1 (TRPA1) Antagonists: Ligand-Based Discovery, Activity in a Rodent Asthma Model, and Mechanism-of-Action via Cryogenic Electron Microscopy.
J.Med.Chem., 64, 2021
|
|
8ILD
 
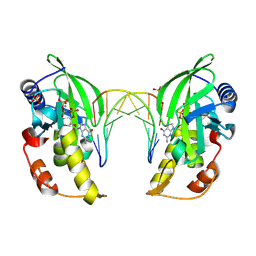 | | The crystal structure of native dGTP:DNApre-I:Pol X substrate ternary complex | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*AP*TP*CP*CP*T)-3'), GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Qin, T, Chen, Y.Q, Gan, J.H, Huang, Z. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insight into Polymerase Mechanism via a Chiral Center Generated with a Single Selenium Atom.
Int J Mol Sci, 24, 2023
|
|
8ILH
 
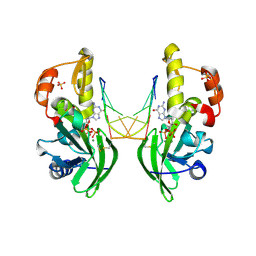 | | The crystal structure of dG(Se-Sp)-DNA:Pol X product binary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOSELENOPHOSPHATE, DNA (5'-D(*CP*GP*GP*AP*TP*CP*C)-3'), MAGNESIUM ION, ... | | Authors: | Qin, T, Gan, J.H, Huang, Z. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insight into Polymerase Mechanism via a Chiral Center Generated with a Single Selenium Atom.
Int J Mol Sci, 24, 2023
|
|
8ILI
 
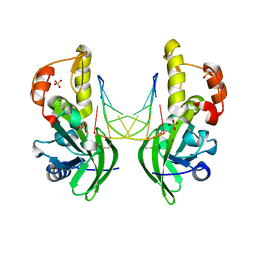 | | The crystal structure of dG(Se-Rp)-DNA:Pol X product binary complex | | Descriptor: | DNA (5'-D(*CP*GP*GP*AP*TP*CP*CP*(7S8))-3'), MAGNESIUM ION, Repair DNA polymerase X, ... | | Authors: | Qin, T, Gan, J.H, Huang, Z. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight into Polymerase Mechanism via a Chiral Center Generated with a Single Selenium Atom.
Int J Mol Sci, 24, 2023
|
|
8ILE
 
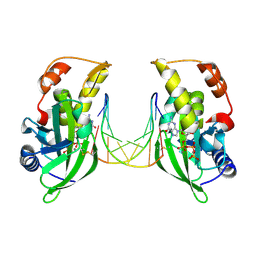 | | The crystal structure of dGTPalphaSe-Rp:DNApre-II:Pol X substrate ternary complex | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*AP*TP*CP*CP*A)-3'), MANGANESE (II) ION, Repair DNA polymerase X, ... | | Authors: | Qin, T, Gan, J.H, Huang, Z. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insight into Polymerase Mechanism via a Chiral Center Generated with a Single Selenium Atom.
Int J Mol Sci, 24, 2023
|
|
8ILG
 
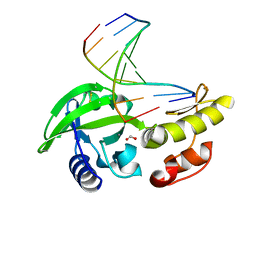 | | The crystal structure of dG-DNA:Pol X product binary complex | | Descriptor: | DNA (5'-D(*CP*GP*GP*AP*TP*CP*CP*G)-3'), FORMIC ACID, MANGANESE (II) ION, ... | | Authors: | Qin, T, Gan, J.H, Huang, Z. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structural Insight into Polymerase Mechanism via a Chiral Center Generated with a Single Selenium Atom.
Int J Mol Sci, 24, 2023
|
|
5GPP
 
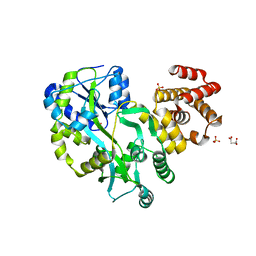 | | Crystal structure of zebrafish ASC PYD domain | | Descriptor: | ACETATE ION, Maltose-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, ... | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2016-08-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of zebrafish ASC.
FEBS J., 285, 2018
|
|
5GPQ
 
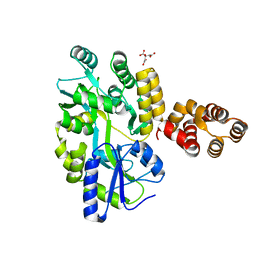 | | Crystal Structure of zebrafish ASC CARD Domain | | Descriptor: | CITRIC ACID, Maltose-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2016-08-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural characterization of zebrafish ASC.
FEBS J., 285, 2018
|
|
8ILF
 
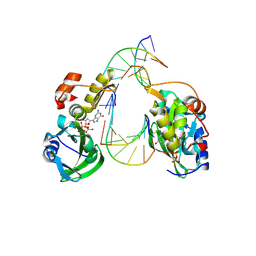 | | The crystal structure of dGTPalphaSe-Sp:DNApre-II:Pol X substrate ternary complex | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*AP*TP*CP*CP*A)-3'), MANGANESE (II) ION, Repair DNA polymerase X, ... | | Authors: | Qin, T, Zhao, Q.W, Gan, J.H, Huang, Z. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insight into Polymerase Mechanism via a Chiral Center Generated with a Single Selenium Atom.
Int J Mol Sci, 24, 2023
|
|
4RFM
 
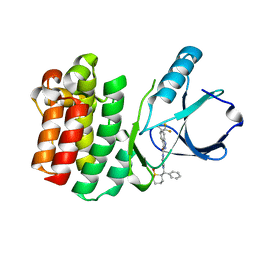 | | ITK kinase domain in complex with compound 1 N-{1-[(1,1-dioxo-1-thian-2-yl)(phenyl)methyl]-1H- pyrazol-4-yl}-5,5-difluoro-5a-methyl-1H,4H,4aH,5H,5aH,6H-cyclopropa[f]indazole-3-carboxamide | | Descriptor: | (4aS,5aR)-N-{1-[(R)-[(2R)-1,1-dioxidotetrahydro-2H-thiopyran-2-yl](phenyl)methyl]-1H-pyrazol-4-yl}-5,5-difluoro-5a-methyl-1,4,4a,5,5a,6-hexahydrocyclopropa[f]indazole-3-carboxamide, Tyrosine-protein kinase ITK/TSK | | Authors: | McEwan, P.A, Barker, J.J, Eigenbrot, C. | | Deposit date: | 2014-09-26 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetrahydroindazoles as Interleukin-2 Inducible T-Cell Kinase Inhibitors. Part II. Second-Generation Analogues with Enhanced Potency, Selectivity, and Pharmacodynamic Modulation in Vivo.
J.Med.Chem., 58, 2015
|
|
6PWB
 
 | | Rigid body fitting of flagellin FlaB, and flagellar coiling proteins, FcpA and FcpB, into a 10 Angstrom structure of the asymmetric flagellar filament purified from Leptospira biflexa Patoc WT cells resolved via subtomogram averaging | | Descriptor: | Flagellar coiling protein A (FcpA), Flagellar coiling protein B (FcpB), Flagellin B1 (FlaB1) | | Authors: | Gibson, K.H, Sindelar, C.V, Trajtenberg, F, Buschiazzo, A, San Martin, F, Mechaly, A. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.83 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the leptospira spirochete.
Elife, 9, 2020
|
|
7Y8W
 
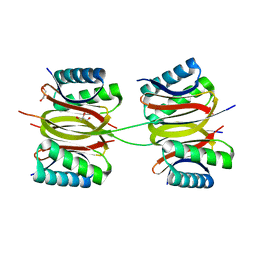 | | Crystal structure of DLC-1/SAO-1 complex | | Descriptor: | Dynein light chain 1, cytoplasmic, GLYCEROL, ... | | Authors: | Yan, H, Zhao, C, Wei, Z, Yu, C. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interaction between DLC-1 and SAO-1 facilitates CED-4 translocation during apoptosis in the Caenorhabditis elegans germline.
Cell Death Discov, 8, 2022
|
|
6H2X
 
 | | MukB coiled-coil elbow from E. coli | | Descriptor: | Chromosome partition protein MukB,Chromosome partition protein MukB | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2018-07-16 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A folded conformation of MukBEF and cohesin.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7DG5
 
 | |
7VTA
 
 | |
7VTB
 
 | |
