6NAY
 
 | |
6NAH
 
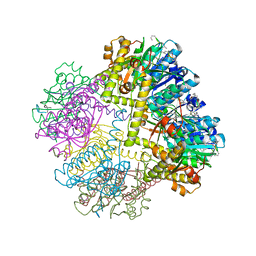 | |
6NAW
 
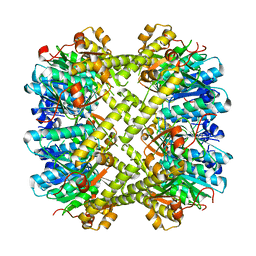 | |
1OVX
 
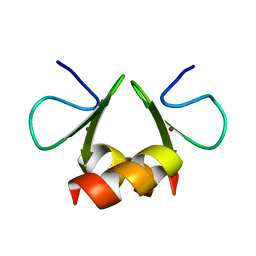 | | NMR structure of the E. coli ClpX chaperone zinc binding domain dimer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, ZINC ION | | Authors: | Donaldson, L.W, Kwan, J, Wojtyra, U, Houry, W.A. | | Deposit date: | 2003-03-27 | | Release date: | 2003-12-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dimeric zinc binding domain of the chaperone ClpX.
J.Biol.Chem., 278, 2003
|
|
2F6I
 
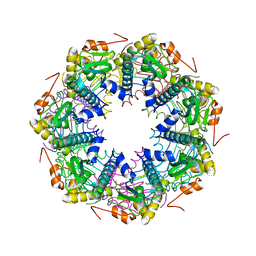 | | Crystal structure of the ClpP protease catalytic domain from Plasmodium falciparum | | Descriptor: | ATP-dependent CLP protease, putative | | Authors: | Mulichak, A, Loppnau, P, Bray, J, Amani, M, Vedadi, M, Wasney, G, Finerty, P, Sundstrom, M, Weigelt, J, Edwards, A, Arrowsmith, C, Hui, R, Plotnikova, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-29 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Clp chaperones and proteases of the human malaria parasite Plasmodium falciparum.
J.Mol.Biol., 404, 2010
|
|
3NBX
 
 | |
6DKF
 
 | |
