1FXT
 
 | | STRUCTURE OF A CONJUGATING ENZYME-UBIQUITIN THIOLESTER COMPLEX | | Descriptor: | UBIQUITIN, UBIQUITIN-CONJUGATING ENZYME E2-24 KDA | | Authors: | Hamilton, K.S, Shaw, G.S, Williams, R.S, Huzil, J.T, McKenna, S, Ptak, C, Glover, M, Ellison, M.J. | | Deposit date: | 2000-09-26 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail.
Structure, 9, 2001
|
|
1J74
 
 | | Crystal Structure of Mms2 | | Descriptor: | MMS2 | | Authors: | Moraes, T.F, Edwards, R.A, McKenna, S, Pastushok, L, Xiao, W, Glover, J.N.M, Ellison, M.J. | | Deposit date: | 2001-05-15 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13.
Nat.Struct.Biol., 8, 2001
|
|
1J7D
 
 | | Crystal Structure of hMms2-hUbc13 | | Descriptor: | MMS2, UBIQUITIN-CONJUGATING ENZYME E2-17 KDA | | Authors: | Moraes, T.F, Edwards, R.A, McKenna, S, Pashushok, L, Xiao, W, Glover, J.N.M, Ellison, M.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-08-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13.
Nat.Struct.Biol., 8, 2001
|
|
1WA1
 
 | | Crystal Structure Of H313Q Mutant Of Alcaligenes Xylosoxidans Nitrite Reductase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Barrett, M.L, Harris, R.L, Antonyuk, S.V, Strange, R.W, Hough, M.A, Eady, R.R, Sawers, G, Hasnain, S.S. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights Into Redox Partner Interactions and Substrate Binding in Nitrite Reductase from Alcaligenes Xylosoxidans: Crystal Structures of the Trp138His and His313Gln Mutants
Biochemistry, 43, 2004
|
|
1WA0
 
 | | Crystal Structure Of W138H Mutant Of Alcaligenes Xylosoxidans Nitrite Reductase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Barrett, M.L, Harris, R.L, Antonyuk, S.V, Strange, R.W, Hough, M.A, Eady, R.R, Sawers, G, Hasnain, S.S. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights Into Redox Partner Interactions and Substrate Binding in Nitrite Reductase from Alcaligenes Xylosoxidans: Crystal Structures of the Trp138His and His313Gln Mutants
Biochemistry, 43, 2004
|
|
1WA2
 
 | | Crystal Structure Of H313Q Mutant Of Alcaligenes Xylosoxidans Nitrite Reductase with nitrite bound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE,, ... | | Authors: | Barrett, M.L, Harris, R.L, Antonyuk, S.V, Strange, R.W, Hough, M.A, Eady, R.R, Sawers, G, Hasnain, S.S. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Insights Into Redox Partner Interactions and Substrate Binding in Nitrite Reductase from Alcaligenes Xylosoxidans: Crystal Structures of the Trp138His and His313Gln Mutants
Biochemistry, 43, 2004
|
|
2YYY
 
 | | Crystal structure of Glyceraldehyde-3-phosphate dehydrogenase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Malay, A.D, Bessho, Y, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of glyceraldehyde-3-phosphate dehydrogenase from the archaeal hyperthermophile Methanocaldococcus jannaschii.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2YXG
 
 | |
2ZGI
 
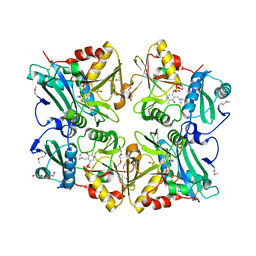 | | Crystal Structure of Putative 4-amino-4-deoxychorismate lyase | | Descriptor: | DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE, Putative 4-amino-4-deoxychorismate lyase, ... | | Authors: | Padmanabhan, B, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-01-22 | | Release date: | 2008-07-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of putative 4-amino-4-deoxychorismate lyase from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
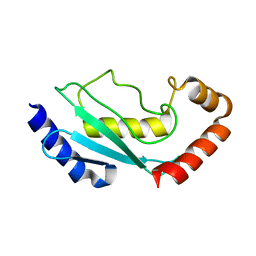
1FZY
 
 | |
