3UXA
 
 | | Designed protein KE59 R1 7/10H | | Descriptor: | Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-05 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UZ5
 
 | | Designed protein KE59 R13 3/11H | | Descriptor: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R13 3/11H, PHOSPHATE ION, ... | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UZJ
 
 | | Designed protein KE59 R13 3/11H with benzotriazole | | Descriptor: | 1H-benzotriazole, Kemp eliminase KE59 R13 3/11H, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY8
 
 | | Designed protein KE59 R5_11/5F | | Descriptor: | Kemp eliminase KE59 R5_11/5F, SULFATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GY0
 
 | |
4IBR
 
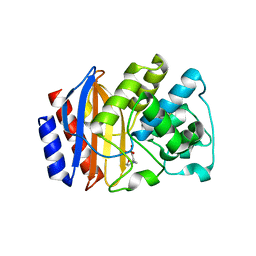 | | Crystal structure of stabilized TEM-1 beta-lactamase variant v.13 carrying G238S/E104K mutations | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, TEM-94 ES-beta-lactamase | | Authors: | Dellus-Gur, E, Toth-Petroczy, A, Elias, M, Tawfik, D.S. | | Deposit date: | 2012-12-09 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | What Makes a Protein Fold Amenable to Functional Innovation? Fold Polarity and Stability Trade-offs.
J.Mol.Biol., 425, 2013
|
|
4IBX
 
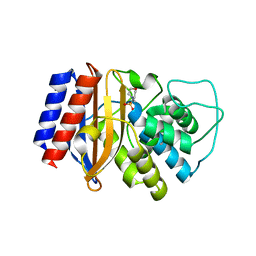 | | Crystal structure of stabilized TEM-1 beta-lactamase variant v.13 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase TEM, CALCIUM ION, ... | | Authors: | Dellus-Gur, E, Toth-Petroczy, A, Elias, M, Tawfik, D.S. | | Deposit date: | 2012-12-09 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | What Makes a Protein Fold Amenable to Functional Innovation? Fold Polarity and Stability Trade-offs.
J.Mol.Biol., 425, 2013
|
|
4GY1
 
 | |
6CJN
 
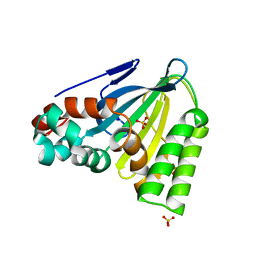 | | Crystal Structure of Chalcone Isomerase from Medicago Sativa with the G95T mutation | | Descriptor: | Chalcone--flavonone isomerase 1, SULFATE ION | | Authors: | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
6CJO
 
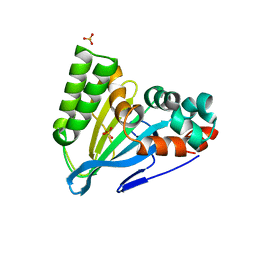 | | Crystal Structure of Chalcone Isomerase from Medicago Sativa with the G95S mutation. | | Descriptor: | Chalcone--flavonone isomerase 1, SULFATE ION | | Authors: | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
3NQ8
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution R4 8/5A | | Descriptor: | BENZAMIDINE, NITRATE ION, deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3NPV
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution | | Descriptor: | deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3NQ2
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution R2 3/5G | | Descriptor: | IMIDAZOLE, deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3NR0
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution R6 6/10A | | Descriptor: | deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3NPX
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution | | Descriptor: | deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3NQV
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution R5 7/4A | | Descriptor: | BENZAMIDINE, deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3NPU
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution | | Descriptor: | deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3NPW
 
 | | In silico designed of an improved Kemp eliminase KE70 mutant by computational design and directed evolution | | Descriptor: | deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3QVV
 
 | |
3QVU
 
 | | Crystal structure of Ancestral variant b9 of SULT 1A1 in complex with PAP and p-nitrophenol | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-3'-5'-DIPHOSPHATE, P-NITROPHENOL, ... | | Authors: | Alcolombri, U, Elias, M, Tawfik, D.S. | | Deposit date: | 2011-02-26 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Directed evolution of sulfotransferases and paraoxonases by ancestral libraries.
J.Mol.Biol., 411, 2011
|
|
3Q2D
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution | | Descriptor: | 5-nitro-1H-benzotriazole, Deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Murphy, P, Dym, O, Albeck, S, Kiss, G, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3SRG
 
 | | Serum paraoxonase-1 by directed evolution at pH 6.5 in complex with 2-hydroxyquinoline | | Descriptor: | BROMIDE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ben David, M, Elias, M, Silman, I, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2011-07-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Catalytic versatility and backups in enzyme active sites: the case of serum paraoxonase 1.
J.Mol.Biol., 418, 2012
|
|
3SRE
 
 | | Serum paraoxonase-1 by directed evolution at pH 6.5 | | Descriptor: | BROMIDE ION, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Ben David, M, Elias, M, Silman, I, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2011-07-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Catalytic versatility and backups in enzyme active sites: the case of serum paraoxonase 1.
J.Mol.Biol., 418, 2012
|
|
6GBJ
 
 | | Repertoires of functionally diverse enzymes through computational design at epistatic active-site positions | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Khersonsky, O, Lipsh, R, Avizemer, Z, Goldsmith, M, Ashani, Y, Leader, H, Dym, O, Rogotner, S, Trudeau, D, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2018-04-15 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Automated Design of Efficient and Functionally Diverse Enzyme Repertoires.
Mol. Cell, 72, 2018
|
|
6GMU
 
 | | Serum paraoxonase-1 by directed evolution with the L69G/H134R/F222S/T332S mutations | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ben-David, M, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2018-05-28 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enzyme Evolution: An Epistatic Ratchet versus a Smooth Reversible Transition.
Mol.Biol.Evol., 37, 2020
|
|
