6SVC
 
 | | Protein allostery of the WW domain at atomic resolution: apo structure | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1A87
 
 | | COLICIN N | | Descriptor: | COLICIN N | | Authors: | Vetter, I.R, Parker, M.W, Tucker, A.D, Lakey, J.H, Pattus, F, Tsernoglou, D. | | Deposit date: | 1998-04-03 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a colicin N fragment suggests a model for toxicity.
Structure, 6, 1998
|
|
2O7Q
 
 | |
1AL1
 
 | | CRYSTAL STRUCTURE OF ALPHA1: IMPLICATIONS FOR PROTEIN DESIGN | | Descriptor: | ALPHA HELIX PEPTIDE: ELLKKLLEELKG, SULFATE ION | | Authors: | Hill, C.P, Anderson, D.H, Wesson, L, Degrado, W.F, Eisenberg, D. | | Deposit date: | 1990-07-02 | | Release date: | 1991-10-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of alpha 1: implications for protein design.
Science, 249, 1990
|
|
4QDN
 
 | | Crystal Structure of the endo-beta-N-acetylglucosaminidase from Thermotoga maritima | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Flagellar protein FlgJ [peptidoglycan hydrolase], PHOSPHATE ION | | Authors: | Lipski, A, Nurizzo, D, Bourne, Y, Vincent, F. | | Deposit date: | 2014-05-14 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical characterization of the beta-N-acetylglucosaminidase from Thermotoga maritima: Toward rationalization of mechanistic knowledge in the GH73 family.
Glycobiology, 25, 2015
|
|
4QI5
 
 | | Dehydrogenase domain of Myriococcum thermophilum cellobiose dehydrogenase with bound cellobionolactam, MtDH | | Descriptor: | (2R,3R,4R,5R)-4,5-dihydroxy-2-(hydroxymethyl)-6-oxopiperidin-3-yl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
1RQP
 
 | | Crystal structure and mechanism of a bacterial fluorinating enzyme | | Descriptor: | 5'-fluoro-5'-deoxyadenosine synthase, S-ADENOSYLMETHIONINE | | Authors: | Dong, C, Huang, F, Deng, H, Schaffrath, C, Spencer, J.B, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2003-12-06 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and mechanism of a bacterial fluorinating enzyme
Nature, 427, 2004
|
|
7XFG
 
 | |
6SC5
 
 | | dAb3/HOIP-RBR-Ligand2 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF31, SULFATE ION, ... | | Authors: | Tsai, Y.-C.I, Johansson, H, House, D, Rittinger, K. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single-Domain Antibodies as Crystallization Chaperones to Enable Structure-Based Inhibitor Development for RBR E3 Ubiquitin Ligases.
Cell Chem Biol, 27, 2020
|
|
1B88
 
 | | V-ALPHA 2.6 MOUSE T CELL RECEPTOR (TCR) DOMAIN | | Descriptor: | T CELL RECEPTOR V-ALPHA DOMAIN | | Authors: | Plaksin, D, Chacko, S, Navaza, J, Margulies, D.H, Padlan, E.A. | | Deposit date: | 1999-02-09 | | Release date: | 1999-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The X-ray crystal structure of a Valpha2.6Jalpha38 mouse T cell receptor domain at 2.5 A resolution: alternate modes of dimerization and crystal packing.
J.Mol.Biol., 289, 1999
|
|
3HVI
 
 | | Rat catechol O-methyltransferase in complex with a catechol-type, N6-ethyladenine-containing bisubstrate inhibitor | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2009-06-16 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Molecular recognition at the active site of catechol-o-methyltransferase: energetically favorable replacement of a water molecule imported by a bisubstrate inhibitor.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
7ZL8
 
 | | NME1 in complex with succinyl-CoA | | Descriptor: | Nucleoside diphosphate kinase A, SUCCINYL-COENZYME A | | Authors: | Garcia-Saez, I, Iuso, D, Khochbin, S, Petosa, C. | | Deposit date: | 2022-04-14 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Nucleoside diphosphate kinases 1 and 2 regulate a protective liver response to a high-fat diet.
Sci Adv, 9, 2023
|
|
7ZLW
 
 | | NME1 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleoside diphosphate kinase A | | Authors: | Garcia-Saez, I, Iuso, D, Khochbin, S, Petosa, C. | | Deposit date: | 2022-04-15 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nucleoside diphosphate kinases 1 and 2 regulate a protective liver response to a high-fat diet.
Sci Adv, 9, 2023
|
|
5FU6
 
 | | NOT module of the human CCR4-NOT complex (Crystallization mutant) | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 2, CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 3 | | Authors: | Raisch, T, Bhandari, D, Sabath, K, Helms, S, Valkov, E, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2016-01-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Distinct Modes of Recruitment of the Ccr4-not Complex by Drosophila and Vertebrate Nanos
Embo J., 35, 2016
|
|
1AYE
 
 | | HUMAN PROCARBOXYPEPTIDASE A2 | | Descriptor: | PROCARBOXYPEPTIDASE A2, ZINC ION | | Authors: | Garcia-Saez, I, Reverte, D, Vendrell, J, Aviles, F.X, Coll, M. | | Deposit date: | 1997-11-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of human procarboxypeptidase A2. Deciphering the basis of the inhibition, activation and intrinsic activity of the zymogen.
EMBO J., 16, 1997
|
|
7ZTK
 
 | | NME1 in complex with CoA | | Descriptor: | COENZYME A, Nucleoside diphosphate kinase A | | Authors: | Garcia-Saez, I, Iuso, D, Khochbin, S, Petosa, C. | | Deposit date: | 2022-05-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nucleoside diphosphate kinases 1 and 2 regulate a protective liver response to a high-fat diet.
Sci Adv, 9, 2023
|
|
8BD3
 
 | | Cryo-EM structure of the Photosystem II - LHCII supercomplex from Chlorella ohadi | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R)-beta,beta-caroten-3-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Fadeeva, M, Klaiman, D, Caspy, I, Nelson, N. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structure of Chlorella ohadii Photosystem II Reveals Protective Mechanisms against Environmental Stress.
Cells, 12, 2023
|
|
5EAA
 
 | | ASPARTATE AMINOTRANSFERASE FROM E. COLI, C191S MUTATION | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-12-29 | | Release date: | 2000-10-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The role of residues outside the active site: structural basis for function of C191 mutants of Escherichia coli aspartate aminotransferase.
Protein Eng., 13, 2000
|
|
5EI0
 
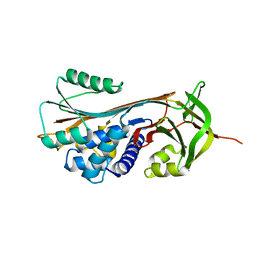 | | Structure of RCL-cleaved vaspin (serpinA12) | | Descriptor: | Serpin A12 | | Authors: | Pippel, J, Kuettner, B.E, Ulbricht, D, Daberger, J, Schultz, S, Heiker, J.T, Strater, N. | | Deposit date: | 2015-10-29 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of cleaved vaspin (serpinA12).
Biol.Chem., 397, 2016
|
|
1B4L
 
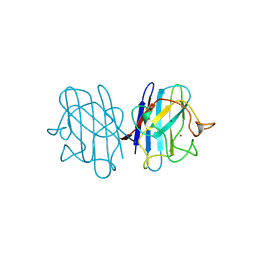 | | 15 ATMOSPHERE OXYGEN YEAST CU/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ZINC ION | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-22 | | Release date: | 1999-12-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1B4T
 
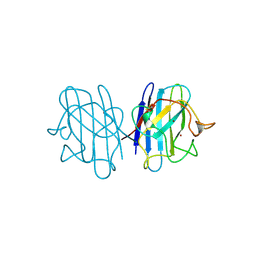 | | H48C YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-23 | | Release date: | 1999-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
4OD2
 
 | |
8CLQ
 
 | |
8CLV
 
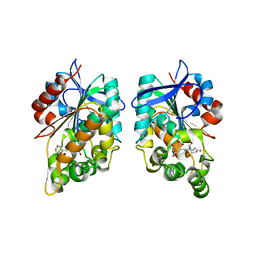 | |
8CPO
 
 | | Crystal structure of the PolB16_OarG intein variant S1A, N183A, C111A, C165A | | Descriptor: | PolB16 Intein Cys-less | | Authors: | Kattelmann, S, Pasch, T, Mootz, H.D, Kuemmel, D. | | Deposit date: | 2023-03-03 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analysis of a novel atypically split intein reveals a conserved histidine specific to cysteine-less inteins.
Chem Sci, 14, 2023
|
|
