8IG9
 
 | |
8IGB
 
 | |
8IG6
 
 | | Crystal structure of MERS main protease in complex with GC376 | | Descriptor: | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, ORF1a | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
5V93
 
 | | Cryo-EM structure of the 70S ribosome from Mycobacterium tuberculosis bound with Capreomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Yang, K, Chang, J.-Y, Cui, Z, Li, X, Meng, R, Duan, L, Thongchol, J, Jakana, J, Huwe, C, Sacchettini, J, Zhang, J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-20 | | Last modified: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis.
Nucleic Acids Res., 45, 2017
|
|
2MLB
 
 | | NMR solution structure of a computational designed protein based on template of human erythrocytic ubiquitin | | Descriptor: | redesigned ubiquitin | | Authors: | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | Deposit date: | 2014-02-21 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
2MN4
 
 | | NMR solution structure of a computational designed protein based on structure template 1cy5 | | Descriptor: | Computational designed protein based on structure template 1cy5 | | Authors: | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | Deposit date: | 2014-03-28 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
2LKZ
 
 | | Solution structure of the second RRM domain of RBM5 | | Descriptor: | RNA-binding protein 5 | | Authors: | Song, Z, Wu, P, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2011-10-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RRM domain of RBM5 and its unusual binding characters for different RNA targets
Biochemistry, 2012
|
|
6VXN
 
 | | Cryo-EM structure of Arabidopsis thaliana MSL1 A320V | | Descriptor: | DODECANE, Mechanosensitive ion channel protein 1, mitochondrial | | Authors: | Deng, Z, Zhang, J, Yuan, P. | | Deposit date: | 2020-02-22 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural mechanism for gating of a eukaryotic mechanosensitive channel of small conductance.
Nat Commun, 11, 2020
|
|
6VXM
 
 | | Cryo-EM structure of Arabidopsis thaliana MSL1 | | Descriptor: | EICOSANE, Mechanosensitive ion channel protein 1, mitochondrial | | Authors: | Deng, Z, Zhang, J, Yuan, P. | | Deposit date: | 2020-02-22 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural mechanism for gating of a eukaryotic mechanosensitive channel of small conductance.
Nat Commun, 11, 2020
|
|
6WDO
 
 | | Cryo-EM structure of mitochondrial calcium uniporter holocomplex in high Ca2+ | | Descriptor: | CALCIUM ION, Calcium uniporter protein, mitochondrial, ... | | Authors: | Feng, L, Zhang, J, Fan, M. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and mechanism of the mitochondrial Ca2+uniporter holocomplex.
Nature, 582, 2020
|
|
7N8X
 
 | | Partial C. difficile TcdB and CSPG4 fragment | | Descriptor: | Chondroitin sulfate proteoglycan 4, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-16 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7N9Q
 
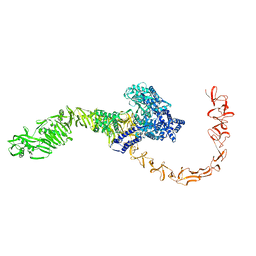 | | State 3 of TcdB and FZD2 at pH5 | | Descriptor: | Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7N95
 
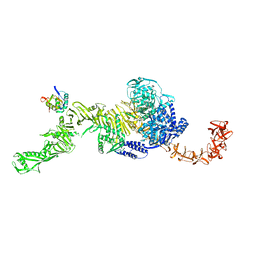 | | state 1 of TcdB and FZD2 at pH5 | | Descriptor: | Frizzled-2, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-16 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7N9R
 
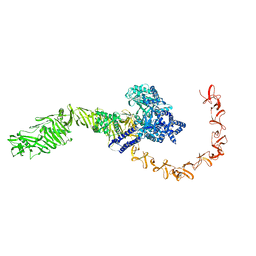 | | state 4 of TcdB and FZD2 at pH5 | | Descriptor: | Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7N9Y
 
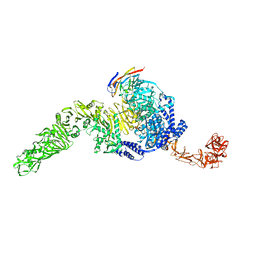 | | Full-length TcdB and CSPG4 (401-560) complex | | Descriptor: | Chondroitin sulfate proteoglycan 4, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7N97
 
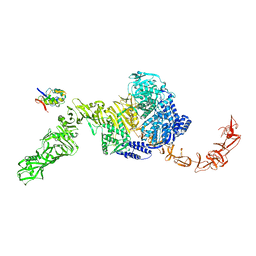 | | State 2 of TcdB and FZD2 at pH5 | | Descriptor: | Frizzled-2, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis for Receptor Recognition of the Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7N9S
 
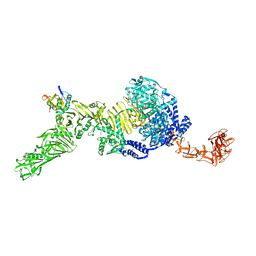 | | TcdB and frizzled-2 CRD complex | | Descriptor: | Frizzled-2, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
6WDN
 
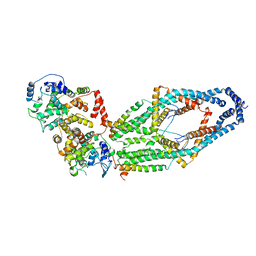 | | Cryo-EM structure of mitochondrial calcium uniporter holocomplex in low Ca2+ | | Descriptor: | Calcium uniporter protein, mitochondrial, Calcium uptake protein 1, ... | | Authors: | Feng, L, Zhang, J, Fan, M. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-27 | | Last modified: | 2020-06-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and mechanism of the mitochondrial Ca2+uniporter holocomplex.
Nature, 582, 2020
|
|
6VXP
 
 | |
5Y4M
 
 | | Discoidin domain of human CASPR2 | | Descriptor: | 1,2-ETHANEDIOL, human CASPR2 Disc domain | | Authors: | Liu, H, Xu, F, Zhang, J, Liang, W. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-08 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural mapping of hot spots within human CASPR2 discoidin domain for autoantibody recognition.
J. Autoimmun., 96, 2019
|
|
8HZR
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, X.Y, Zhang, J, Li, J. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
S46F mutant in complex with PF07321332
To Be Published
|
|
8IU2
 
 | | Cryo-EM structure of Long-wave-sensitive opsin 1 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Peng, Q, Cheng, X.Y, Li, J, Lu, Q.Y, Li, Y.Y, Zhang, J. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of Long-wave-sensitive opsin 1
To Be Published
|
|
8INZ
 
 | | Cryo-EM structure of human HCN3 channel in apo state | | Descriptor: | 4-[[(2~{S},4~{a}~{R},6~{S},8~{a}~{S})-6-[(4~{S},5~{R})-4-[(2~{S})-butan-2-yl]-5,9-dimethyl-decyl]-4~{a}-methyl-2,3,4,5,6,7,8,8~{a}-octahydro-1~{H}-naphthalen-2-yl]oxy]-4-oxidanylidene-butanoic acid, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | | Authors: | Yu, B, Lu, Q.Y, Li, J, Zhang, J. | | Deposit date: | 2023-03-10 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM structure of human HCN3 channel in apo state
To Be Published
|
|
8J39
 
 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
V186F mutant in complex with PF00835231
To Be Published
|
|
8J35
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
G15S mutant in complex with PF00835231
To Be Published
|
|
