1C6C
 
 | |
1C6G
 
 | |
1C6Q
 
 | |
1C6H
 
 | |
1C6I
 
 | |
1C6T
 
 | |
1C6J
 
 | |
1C6M
 
 | |
1C6K
 
 | |
1C6L
 
 | |
1C66
 
 | |
1C6N
 
 | |
1C6A
 
 | |
1L96
 
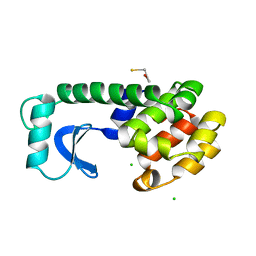 | | STRUCTURE OF A HINGE-BENDING BACTERIOPHAGE T4 LYSOZYME MUTANT, ILE3-> PRO | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Dixon, M, Shewchuk, L, Matthews, B.W. | | Deposit date: | 1992-02-11 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a hinge-bending bacteriophage T4 lysozyme mutant, Ile3-->Pro.
J.Mol.Biol., 227, 1992
|
|
1L97
 
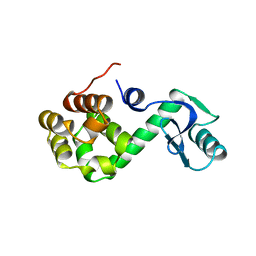 | |
201L
 
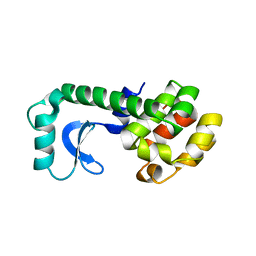 | |
234L
 
 | | T4 LYSOZYME MUTANT M106L | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Lipscomb, L.A, Drew, D.L, Gassner, N, Baase, W.A, Matthews, B.W. | | Deposit date: | 1997-10-07 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Context-dependent protein stabilization by methionine-to-leucine substitution shown in T4 lysozyme.
Protein Sci., 7, 1998
|
|
230L
 
 | | T4 LYSOZYME MUTANT M6L | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Lipscomb, L.A, Gassner, N.C, Snow, S, Eldridge, A.M, Drew, D.L, Baase, W.A, Matthews, B.W. | | Deposit date: | 1997-10-02 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Context-dependent protein stabilization by methionine-to-leucine substitution shown in T4 lysozyme.
Protein Sci., 7, 1998
|
|
256L
 
 | | BACTERIOPHAGE T4 LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Faber, H.R, Matthews, B.W. | | Deposit date: | 1998-02-24 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A mutant T4 lysozyme displays five different crystal conformations.
Nature, 348, 1990
|
|
2ANV
 
 | | crystal structure of P22 lysozyme mutant L86M | | Descriptor: | CHLORIDE ION, IODIDE ION, Lysozyme, ... | | Authors: | Mooers, B.H, Matthews, B.W. | | Deposit date: | 2005-08-11 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Extension to 2268 atoms of direct methods in the ab initio determination of the unknown structure of bacteriophage P22 lysozyme.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2ANX
 
 | |
2B3K
 
 | | Crystal structure of Human Methionine Aminopeptidase Type I in the holo form | | Descriptor: | ACETATE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Addlagatta, A, Hu, X, Liu, J.O, Matthews, B.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for the Functional Differences between Type I and Type II Human Methionine Aminopeptidases(,).
Biochemistry, 44, 2005
|
|
2B3L
 
 | | Crystal structure of type I human methionine aminopeptidase in the apo form | | Descriptor: | ACETIC ACID, GLYCEROL, Methionine aminopeptidase 1, ... | | Authors: | Addlagatta, A, Hu, X, Liu, J.O, Matthews, B.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Functional Differences between Type I and Type II Human Methionine Aminopeptidases(,).
Biochemistry, 44, 2005
|
|
2B3H
 
 | | Crystal structure of Human Methionine Aminopeptidase Type I with a third cobalt in the active site | | Descriptor: | CHLORIDE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Addlagatta, A, Hu, X, Liu, J.O, Matthews, B.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Basis for the Functional Differences between Type I and Type II Human Methionine Aminopeptidases(,).
Biochemistry, 44, 2005
|
|
2B6W
 
 | | T4 Lysozyme mutant L99A at 200 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
